













| General Information: |
|
| Name(s) found: |
MDAR4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 488 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisomal membrane
[IDA]
chloroplast envelope [IDA] |
| Biological Process: |
hydrogen peroxide catabolic process
[IMP][TAS]
|
| Molecular Function: |
monodehydroascorbate reductase (NADH) activity
[IDA][IMP]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGRAFVYVIL GGGVAAGYAA LEFTRRGVSD GELCIISEEP VAPYERPALS KGFLLPEAPA 60
61 RLPSFHTCVG ANDEKLTPKW YKDHGIELVL GTRVKSVDVR RKTLLSSTGE TISYKFLIIA 120
121 TGARALKLEE FGVEGSDAEN VCYLRDLADA NRLATVIQSS SNGNAVVIGG GYIGMECAAS 180
181 LVINKINVTM VFPEAHCMAR LFTPKIASLY EDYYRAKGVK FIKGTVLTSF EFDSNKKVTA 240
241 VNLKDGSHLP ADLVVVGIGI RPNTSLFEGQ LTIEKGGIKV NSRMQSSDSS VYAIGDVATF 300
301 PVKLFGEMRR LEHVDSARKS ARHAVSAIMD PIKTGDFDYL PFFYSRVFAF SWQFYGDPTG 360
361 DVVHFGEYED GKSFGAYWVK KGHLVGSFLE GGTKEEYETI SKATQLKPAV TIDLEELERE 420
421 GLGFAHTVVS QQKVPEVKDI PSAEMVKQSA SVVMIKKPLY VWHAATGVVV AASVAAFAFW 480
481 YGRRRRRW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
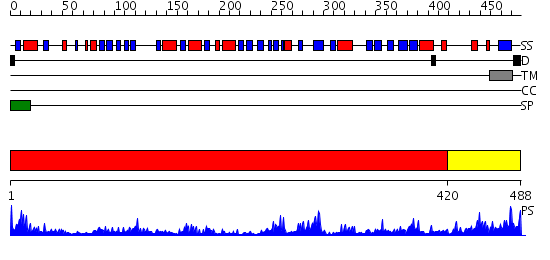
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..419] | 64.522879 | Crystal Structure of Putidaredoxin Reductase from Pseudomonas putida | |
| 2 | View Details | [420..488] | 1.686988 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|