













| General Information: |
|
| Name(s) found: |
DNMT3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1404 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
DNA methylation
[ISS]
embryonic development ending in seed dormancy [IMP] |
| Molecular Function: |
DNA (cytosine-5-)-methyltransferase activity
[IEA][ISS]
DNA binding [IEA] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTKAGKQKK RSVDSDDDVS RERRPKRATS GTNFKEKSLR FSEKYETVEA KKEQIVGDDE 60
61 KEEKGVRFQS FGRVENWTIS GYEDGSPVIW ISTVIADYDC RKPSKKYKKL YDYFFEKACA 120
121 CVEVCKNLST NPDTSLKELL AAVVRSMNGR KIFSSGGVIQ EFVISQGEFI YNQLAGLDET 180
181 SKNHETKFVD NRVLVSLRDE SRKIHKAFSN VALRIDESKV LTSDQLMDGG EDEDLKYAKL 240
241 LQEEEHMKSM DRSRNKRSST TSAPNKFYIK INEDEIAHDY PLPSYYKNTK DETDELVLFN 300
301 AGYAVDARNL PCRTLHNWAL YNSDLMLISL EFLPMKPCAD IDVTYLGQIK EWKIDFGEDM 360
361 IFVLLRTDMA WYRLGKPSEQ YAPWFEPILK TVRIGTSILA LLKNETRMAK LSYTDVIKRL 420
421 CGLEENDQAY ISSTFFDVER YVIVHGQIIL QFLTECPDEY IKRCPFVTGL ASKMQDRHHT 480
481 KWIIKKKRKM LQKGENLNLR RGKAPKVSKM KAMQATTTRL INRIWGEFYS IYSPEDPLEE 540
541 IGAEEEFEEV EDVEEEDENE EEDTIQKAIE VQKADTLKKI RGSCKEMEIR WEGEILGETC 600
601 AGEPLYGQAL VGGRKMDVGG AVILEVDDQG ETPLIYFVEY MFESSDNSKK LHGKLLQRGS 660
661 ETVLGTAANE RELFLTNECL TVQLKDIKGT VSFEIRSRPW GHQYKKEHMA ADKLDRARAE 720
721 ERKAKDLPIE YYCKSLYSPE KGGFFSLPRS DMGLGSGFCS SCKIRENEEE RSKTKLNDSK 780
781 TRFLSNGIKY SVGDFVYQIP NYLSKDRGKR RPVFKYGRNV GLRAFVVCQI LDIVDLKEPK 840
841 KGNTTSFEVK VRRFYRPDDV SAEEAYASDI QEVYYSEDTY ILPPEAIKGK CEVMKKTDMP 900
901 LCREYPILDH VYFCDRFYDS SNGCLKKLPY NMMLKFSTIK DDTLLREKKT ETGSAMLLKP 960
961 DEVPKGKRLA TLDIFAGCGG LSYGLEKAGV SDTKWAIEYE EPAAQAFKQN HPKTTVFVDN1020
1021 CNVILRISWL RLLINDRAIM EKCGDVDDCI STTEAAELAT KLDENQKSTL PLPGQVDFIS1080
1081 GGPPCQGFSR LNRFSDGSWS KNQCQMILAF LSFADYFRPK YFLLENVKTF VSFNEGHTFH1140
1141 LTVASLLEMG YQVRFGLLEA GAYGISQPRK RAFIWAAAPN EVLPEWPEPM HVFNNPGFKI1200
1201 PLSQGLHYAA VQSTKFGAPF RSITVRDAIG DLPPIESGES KINKEEMRGS MTVLTDHICK1260
1261 KMNELNLIRC KKIPKTPGAD WRDLPDEHVN LSNGIVKNIV PNLLNKAKDH NGYKGLYGRL1320
1321 DWHGNLPTCI TNLQPMGLVG MCFHPDQDRI ISVRECARSQ GFPDSYKFSG NIKDKHRQVG1380
1381 NAVPPPLAFA LGRKLKEALH LRNI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
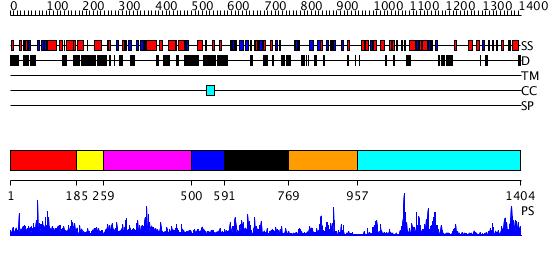
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | 1.004975 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [185..258] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [259..499] | 3.69897 | No description for 2grcA was found. | |
| 4 | View Details | [500..590] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [591..768] | 36.39794 | Crystal structure of the proximal BAH domain of polybromo | |
| 6 | View Details | [769..956] | 3.16899 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [957..1404] | 49.39794 | DNA methylase HaeIII |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)