













| General Information: |
|
| Name(s) found: |
DNMT4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1519 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
DNA mediated transformation
[IMP]
DNA methylation [ISS] |
| Molecular Function: |
DNA (cytosine-5-)-methyltransferase activity
[IEA][ISS]
DNA binding [IEA] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEMETKAGKQ KKRSVDSDDD VSKERRPKRA AACTNFKEKS LRISDKSETV EAKKEQILAE 60
61 EIVAIQLTSS LESNDDPRPN RRLTDFVLHD SEGVPQPVEM LELGDIFIEG VVLPLGDEKK 120
121 EEKGVRFQSF GRVENWNISG YEDGSPVIWI STALADYDCR KPSKKYKKLY DYFFEKACAC 180
181 VEVFKSLSKN PDTSLDELLA AVSRSMSGSK IFSSGGAIQE FVISQGEFIY NQLAGLDETA 240
241 KNHETCFVEN RVLVSLRDHE SNKIHKALSN VALRIDESKV VTSDHLVDGA EDEDVKYAKL 300
301 IQEEEYRKSM ERSRNKRSST TSGGSSRFYI KISEDEIADD YPLPSYYKNT KEETDELVLF 360
361 EAGYEVDTRD LPCRTLHNWT LYNSDSRMIS LEVLPMRPCA EIDVTVFGSG VVAEDDGSGF 420
421 CLDDSESSTS TQSNDHDGMN IFLSQIKEWM IEFGAEMIFV TLRTDMAWYR LGKPSKQYAP 480
481 WFGTVMKTVR VGISIFNMLM RESRVAKLSY ANVIKRLCGL EENDKAYISS KLLDVERYVV 540
541 VHGQIILQLF EEYPDKDIKR CPFVTSLASK MQDIHHTKWI IKKKKKILQK GKNLNPRAGI 600
601 APVVSRMKAM QATTTRLVNR IWGEFYSIYS PEVPSEAINA ENVEEEELEE VEEEDENEED 660
661 DPEENELEAV EIQNSPTPKK IKGISEDMEI KWDGEILGKT SAGEPLYGRA FVGGDVVVVG 720
721 SAVILEVDDQ DDTQLICFVE FMFESSNHSK MLHGKLLQRG SETVLGMAAN ERELFLTNEC 780
781 LTVQLKDIKG TVSLEIRSRL WGHQYRKENI DVDKLDRARA EERKTNGLPT DYYCKSLYSP 840
841 ERGGFFSLPR NDMGLGSGFC SSCKIRENEE ERSKTKLNDS KTGFLSNGIE YHNGDFVYVL 900
901 PNYITKDGLK KGSRRTTLKC GRNVGLKAFV VCQLLDVIVL EESRKASKAS FQVKLTRFYR 960
961 PEDISEEKAY ASDIQELYYS QDTYILPPEA IQGKCEVRKK SDMPLCREYP ILDHIFFCEV1020
1021 FYDSSTGYLK QFPANMKLKF STIKDETLLR EKKGKGVETG TSSGMLMKPD EVPKEKPLAT1080
1081 LDIFAGCGGL SHGLENAGVS TTKWAIEYEE PAGHAFKQNH PEATVFVDNC NVILRAIMEK1140
1141 CGDVDDCVST VEAAELAAKL DENQKSTLPL PGQVDFINGG PPCQGFSGMN RFSHGSWSKV1200
1201 QCEMILAFLS FADYFRPKYF LLENVKKFVT YNKGRTFQLT MASLLEMGYQ VRFGILEAGT1260
1261 YGVSQPRKRV IIWAASPEEV LPEWPEPMHV FDNPGSKISL PRGLRYDAGC NTKFGAPFRS1320
1321 ITVRDTIGDL PPVENGESKI NKEYGTTPAS WFQKKIRGNM SVLTDHICKG LNELNLIRCK1380
1381 KIPKRPGADW RDLPDENVTL SNGLVEKLRP LALSKTAKNH NEWKGLYGRL DWQGNLPISI1440
1441 TDPQPMGKVG MCFHPEQDRI ITVRECARSQ GFPDSYEFSG TTKHKHRQIG NAVPPPLAFA1500
1501 LGRKLKEALY LKSSLQHQS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
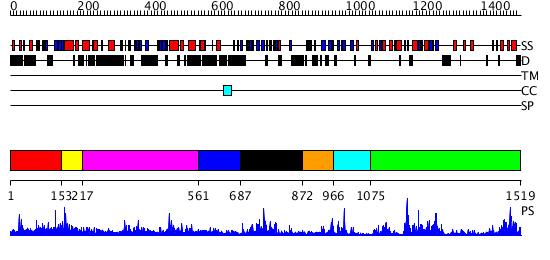
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | 1.01297 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [153..216] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [217..560] | 11.0 | No description for 2r0vA was found. | |
| 4 | View Details | [561..686] | 8.69897 | TAFII250 double bromodomain module | |
| 5 | View Details | [687..871] | 34.69897 | Crystal structure of the proximal BAH domain of polybromo | |
| 6 | View Details | [872..965] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [966..1074] | 2.223995 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1075..1519] | 39.154902 | DNA methylase HaeIII |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)