













| General Information: |
|
| Name(s) found: |
SOT18_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 350 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
glucosinolate biosynthetic process
[IDA]
|
| Molecular Function: |
indol-3-yl-methyl-desulfoglucosinolate sulfotransferase activity
[IDA]
3-methylthiopropyl-desulfoglucosinolate sulfotransferase activity [IDA] desulfoglucosinolate sulfotransferase activity [IDA] 4-methylthiobutyl-desulfoglucosinolate sulfotransferase activity [IDA] 5-methylthiopentyl-desulfoglucosinolate sulfotransferase activity [IDA] 7-methylthioheptyl-desulfoglucosinolate sulfotransferase activity [IDA] sulfotransferase activity [ISS] 8-methylthiooctyl-desulfoglucosinolate sulfotransferase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESETLTAKA TITTTTLPSH DETKTESTEF EKNQKRYQDL ISTFPHEKGW RPKEPLIEYG 60
61 GYWWLPSLLE GCIHAQEFFQ ARPSDFLVCS YPKTGTTWLK ALTFAIANRS RFDDSSNPLL 120
121 KRNPHEFVPY IEIDFPFFPE VDVLKDKGNT LFSTHIPYEL LPDSVVKSGC KMVYIWREPK 180
181 DTFISMWTFL HKERTELGPV SNLEESFDMF CRGLSGYGPY LNHILAYWKA YQENPDRILF 240
241 LKYETMRADP LPYVKSLAEF MGHGFTAEEE EKGVVEKVVN LCSFETLKNL EANKGEKDRE 300
301 DRPGVYANSA YFRKGKVGDW SNYLTPEMAA RIDGLMEEKF KGTGLLEHGK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
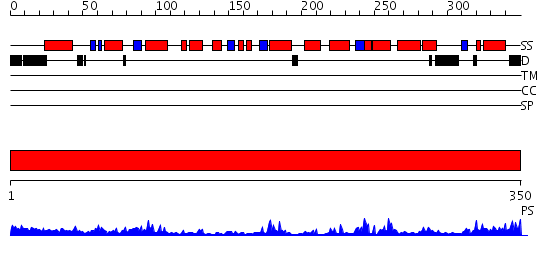
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..350] | 59.39794 | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulphotransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)