













| General Information: |
|
| Name(s) found: |
gi|8886954
[NCBI NR]
gi|51969260 [NCBI NR] gi|29028862 [NCBI NR] gi|25317943 [NCBI NR] gi|18394881 [NCBI NR] gi|110743122 [NCBI NR] |
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 246 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[ISS]
|
| Biological Process: |
protein amino acid glycosylation
[ISS]
|
| Molecular Function: |
dolichyl-phosphate beta-D-mannosyltransferase activity
[ISS]
elongation factor-2 kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADEMETKGE KKYKYSIIIP TYNERLNIAI IVYLIFKHLR DVDFEIIVVD DGSPDGTQEI 60
61 VKQLQQLYGE DRILLRARAK KLGLGTAYIH GLKHATGDFV VIMDADLSHH PKYLPSFIKK 120
121 QLETNASIVT GTRYVKGGGV HGWNLMRKLT SRGANVLAQT LLWPGVSDLT GSFRLYKKSA 180
181 LEDVISSCVS KGYVFQMEMI VRATRKGYHI EEVPITFVDR VFGTSKLGGS EIVEYLKGLV 240
241 YLLLTT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
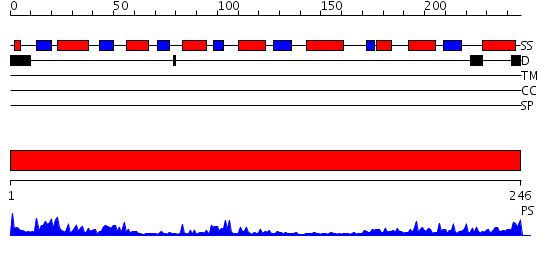
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..246] | 29.154902 | The Crystal Structure of UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase-T1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)