













| General Information: |
|
| Name(s) found: |
FAO4B_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 748 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
oxidoreductase activity, acting on CH-OH group of donors
[IEA]
FAD binding [IEA] oxidoreductase activity [IEA] electron carrier activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDVRRRNRG HPLLRSKKRG EGYNHGFSPS QIQSLAVICQ TFLPPETTSE QQAVNSFHVA 60
61 SSTQPPFTDE VAEMIVKNGR SEAVKVLRII LMILSFRFGT LLLCGSLCLD KSWPFVLKFS 120
121 QLPLDKREAI LRNWSRQSGF LLPFRITFFL AKFYTLFYFF SQTDENLKNP ALEAIGYCID 180
181 GTERSSNKKS EADEKRRPLE KGIIETMHES DVTITQSLTE KGVHVARDDG DNVYRIRCDA 240
241 VVVGSGSGGG VAAANLAKAG LKVLVLEKGN YFTAHDYSGL EVPSMLELYE KGGLLTTVDG 300
301 KFMLLAGSAV GGGTAVNWSA SIRTPDHVLQ EWSEGSKIKF FGSQEYQSAM DEVTIRIGVT 360
361 ERCVKHGFQN QVLRKGCERL GLQVESVPRN SPEDHYCGLC GYGCRAGAKN GTDQTWLVDA 420
421 VENGAVILTG IKAERFVLVD NTSSSNERKK RCVGVFASSV GGKIGKKFII EARVTVSSAG 480
481 SLLTPPLMLS SGLKNPNIGR NLKLHPVLMT WGYFPEKDSE FSGKMYEGGI ITSVHHMNDT 540
541 ESGCKAILEN PLIGPASYAG LSPWVSGPDL KERMIKYGRT AHLFALVRDL GSGEVMMENE 600
601 VTYRTTKKDR ENLRAGLRQA LRVSVAAGAV EVGTYRSDGQ KMKCEAITKE AMEEFLDEVD 660
661 AVGGVGTKGE YWTTYFSAHQ MGSCRMGVTA EEGALDENGE SWEAEGLFVC DGSILPSAVG 720
721 VNPMITIQST AYCISSKIVD SLQNKTKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
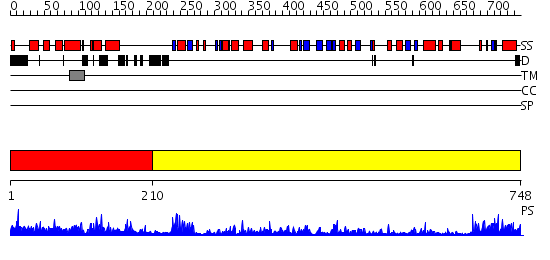
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..209] | 1.069963 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [210..748] | 67.0 | Hydroxynitrile lyase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|