













| General Information: |
|
| Name(s) found: |
APS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 463 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] plasma membrane [IDA] |
| Biological Process: |
response to cadmium ion
[IEP]
|
| Molecular Function: |
sulfate adenylyltransferase (ATP) activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASMAAVLSK TPFLSQPLTK SSPNSDLPFA AVSFPSKSLR RRVGSIRAGL IAPDGGKLVE 60
61 LIVEEPKRRE KKHEAADLPR VELTAIDLQW MHVLSEGWAS PLGGFMRESE FLQTLHFNSL 120
121 RLDDGSVVNM SVPIVLAIDD EQKARIGEST RVALFNSDGN PVAILSDIEI YKHPKEERIA 180
181 RTWGTTAPGL PYVDEAITNA GNWLIGGDLE VLEPVKYNDG LDRFRLSPAE LRKELEKRNA 240
241 DAVFAFQLRN PVHNGHALLM TDTRRRLLEM GYKNPILLLH PLGGFTKADD VPLDWRMKQH 300
301 EKVLEDGVLD PETTVVSIFP SPMHYAGPTE VQWHAKARIN AGANFYIVGR DPAGMGHPVE 360
361 KRDLYDADHG KKVLSMAPGL ERLNILPFRV AAYDKTQGKM AFFDPSRPQD FLFISGTKMR 420
421 TLAKNNENPP DGFMCPGGWK VLVDYYESLT PAGNGRLPEV VPV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
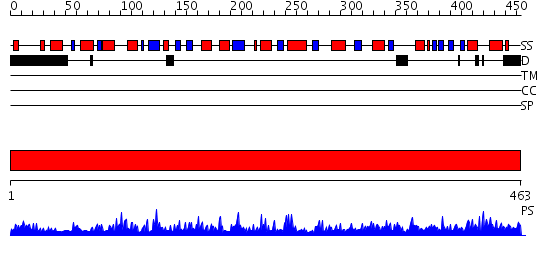
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..463] | 177.0 | The crystal structure of human 3'-phosphoadenosine-5'-phosphosulfate synthetase 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)