













| General Information: |
|
| Name(s) found: |
DSK2B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 551 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein modification process
[IEA]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGEGDSSQP QSGEGEAVAV NIRCSNGTKF SVKTSLDSTV ESFKELVAQS SDVPANQQRL 60
61 IYKGRILKDD QTLLSYGLQA DHTIHMVRGS APSSAPPPAP AASQTTAPSV TRGVGSDNSS 120
121 NLGGASPGES LFPGLGFNPL GGGNAMSGLF GAGLPDLVQT QQQLAQNPNM IRDMMNTPAI 180
181 QNLMNNPEFM RSMIMNNPQM RELVDRNPEL GHVLNDPSIL RQTLEAARNP ELMREMMRNT 240
241 DRAMSNIESM PEGFNMLRRM YENVQEPLMN ATTMSGNAGN NTGSNPFAAL LGNQGVTTQG 300
301 SDASNNSSTP NAGTGTIPNA NPLPNPWGAT GGQTTAPGRT NVGGDARSPG LGGLGGLGSL 360
361 GGLGGLGMLG ADSPLGATPD ASQLSQLLQN PAISQMMQSV FSNPQYMNQL MSLNPQLRSM 420
421 LDSNPQLREM MQNPDFLRQF SSPEMMQQMM TLQQSLSQNR NTASQDAGQT GAATGNNGGL 480
481 DLLMNMFGSL GAGGLSGTNQ SNVPPEERYA TQLQQLQEMG FYDRAENIRA LLATNGNVNA 540
541 AVERLLGSIG Q |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
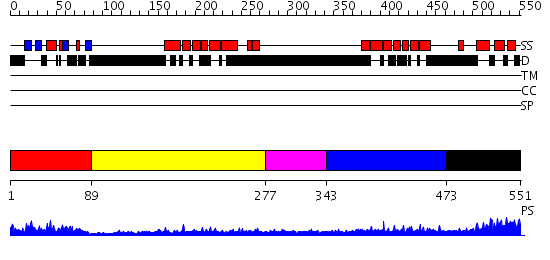
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | 20.0 | Solution Structure of S5a UIM-1/Ubiquitin Complex | |
| 2 | View Details | [89..276] | 1.01 | Structure of the DNA repair protein hHR23a | |
| 3 | View Details | [277..342] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [343..472] | 5.522879 | Sec24 | |
| 5 | View Details | [473..551] | 8.09691 | Solution Structure of RSGI RUH-016, a UBA Domain from mouse cDNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)