













| General Information: |
|
| Name(s) found: |
SUS3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
sucrose biosynthetic process
[ISS]
biosynthetic process [IEA] sucrose metabolic process [IEA] |
| Molecular Function: |
UDP-glycosyltransferase activity
[ISS]
transferase activity, transferring glycosyl groups [ISS] sucrose synthase activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANPKLTRVL STRDRVQDTL SAHRNELVAL LSRYVDQGKG ILQPHNLIDE LESVIGDDET 60
61 KKSLSDGPFG EILKSAMEAI VVPPFVALAV RPRPGVWEYV RVNVFELSVE QLTVSEYLRF 120
121 KEELVDGPNS DPFCLELDFE PFNANVPRPS RSSSIGNGVQ FLNRHLSSVM FRNKDCLEPL 180
181 LDFLRVHKYK GHPLMLNDRI QSISRLQIQL SKAEDHISKL SQETPFSEFE YALQGMGFEK 240
241 GWGDTAGRVL EMMHLLSDIL QAPDPSSLEK FLGMVPMVFN VVILSPHGYF GQANVLGLPD 300
301 TGGQVVYILD QVRALETEML LRIKRQGLDI SPSILIVTRL IPDAKGTTCN QRLERVSGTE 360
361 HTHILRVPFR SEKGILRKWI SRFDVWPYLE NYAQDAASEI VGELQGVPDF IIGNYSDGNL 420
421 VASLMAHRMG VTQCTIAHAL EKTKYPDSDI YWKDFDNKYH FSCQFTADLI AMNNADFIIT 480
481 STYQEIAGTK NTVGQYESHG AFTLPGLYRV VHGIDVFDPK FNIVSPGADM TIYFPYSEET 540
541 RRLTALHGSI EEMLYSPDQT DEHVGTLSDR SKPILFSMAR LDKVKNISGL VEMYSKNTKL 600
601 RELVNLVVIA GNIDVNKSKD REEIVEIEKM HNLMKNYKLD GQFRWITAQT NRARNGELYR 660
661 YIADTRGAFA QPAFYEAFGL TVVEAMTCGL PTFATCHGGP AEIIEHGLSG FHIDPYHPEQ 720
721 AGNIMADFFE RCKEDPNHWK KVSDAGLQRI YERYTWKIYS ERLMTLAGVY GFWKYVSKLE 780
781 RRETRRYLEM FYILKFRDLV KTVPSTADD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
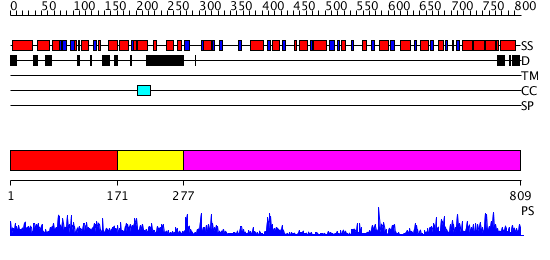
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..170] | 2.031941 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [171..276] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [277..809] | 61.39794 | No description for 2r60A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)