













| General Information: |
|
| Name(s) found: |
CLPX1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 579 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA]
mitochondrion [IDA] |
| Biological Process: |
protein transport
[IEA]
|
| Molecular Function: |
ATPase activity
[IEA][ISS]
nucleotide binding [IEA] ATP binding [IEA] protein binding [IEA] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAALRSNTS RETASLTLSH FRYFIFNRIH TARTATSPPH CNHRSKSDEK FPYKISSLGT 60
61 SFLDNRGGGE RRNSTKCYAA QKKLSGVGSS VVILSSQGDP PDLWQPPGDG VSVRVNGSSV 120
121 NLGRGGGGGG SSPGGPGNGT GSNSKEDCWG GSNLGSDFPT PKEICKGLNK FVIGQERAKK 180
181 VLSVAVYNHY KRIYHESSQK RSAGETDSTA AKPADDDMVE LEKSNILLMG PTGSGKTLLA 240
241 KTLARFVNVP FVIADATTLT QAGYVGEDVE SILYKLLTVA DYNVAAAQQG IVYIDEVDKI 300
301 TKKAESLNIS RDVSGEGVQQ ALLKMLEGTI VNVPEKGARK HPRGDNIQID TKDILFICGG 360
361 AFVDIEKTIS ERRHDSSIGF GAPVRANMRA GGVTNAAVAS NLMETVESSD LIAYGLIPEF 420
421 VGRFPVLVSL SALTENQLMQ VLTEPKNALG KQYKKMYQMN SVKLHFTESA LRLIARKAIT 480
481 KNTGARGLRA LLESILMDSM YEIPDEGTGS DMIEAVVVDE EAVEGEGRRG SGAKILRGKG 540
541 ALARYLSETN SKDSPQTTKE GSDGETEVEA EIPSVVASM |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
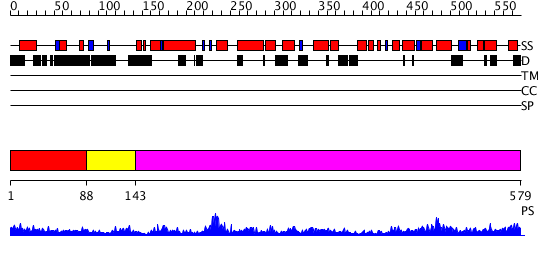
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 2.402996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [88..142] | 4.0 | NMR structure of the E. coli ClpX chaperone zinc binding domain dimer | |
| 3 | View Details | [143..579] | 69.0 | Crystal structure of helicobacter pylori ClpX |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)