













| General Information: |
|
| Name(s) found: |
MMT1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1071 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
S-adenosylmethionine metabolic process
[IMP]
|
| Molecular Function: |
S-adenosylmethionine-dependent methyltransferase activity
[IMP][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADLSSVDEF LNQCKQSGDA AYGALRSVLE RLEDPNTRSK ARIFLSDIYK RVGSSETSLQ 60
61 TYHFHIQDIY LDQYEGFQSR KKLTMMVIPS IFIPEDWSFT FYEGLNRHPD TIFKDKTVSE 120
121 LGCGNGWISI AIAAKWLPSK VYGLDINPRA VKISWINLYL NALDDNGEPV YDEEKKTLLD 180
181 RVEFYESDLL GYCRDNKIQL ERIVGCIPQI LNPNPEAMSK LITENASEEF LHSLSNYCAL 240
241 QGFVEDQFGL GLIARAVEEG ISVIKPAGIM IFNMGGRPGQ GVCRRLFERR GVRVTQMWQT 300
301 KILQAADTDI SALVEIERSS PHRFEFFMGL SGDQPICART AWAYGKAGGR ISHALSVYSC 360
361 QIRQPNLVKI IFDFLKNGFQ EISNSLDLSF EDETVADEKI PFLAYLASVL KNSSYFPFEP 420
421 PAGSKRFCSL IAGFMRTYHR IPINQDNIVV FPSRAVAIES AFRLFSPRLA IVDEHLTRQL 480
481 PRSWLTSLAI EDTSMDKSDD QITVIESPHQ SDLMIELIKK LKPQVVVTGM APFEVITSSS 540
541 FLHLLEVTKE IGCRLFLDIS DHFELSSLPA SNGVLKYLAE NQLPSHAAII CGLVKNKVYS 600
601 DLEVAFVITE VDAIAKALSK TVEVLEGHTA IISQYYYGCL FHELLAFQLA DRHAPAERES 660
661 EKAKSEEIIG FSSSAVSILK DAELSVTEID ETSLIHMDVD QSFLQIPQSV KAAIFESFVR 720
721 QNISEAEVDI NPSIKQFVWS NYGFPTKSST GFVYADGSLA LFNKLVICCA QEGGTLCLPA 780
781 GTNGNYVAAA KFLKANVVNI PTESSDGFKL TEKTLTKALE SVKKPWVCIS GPTVSPTGLV 840
841 YSNEEMDILL STCAKFGAKV IIDTSFSGLE YSATSWDLKN ALSKMDSSLS VSLLGCLSLN 900
901 LLSGAIKLGF LVLDQSLIDA FHTLPGLSKP HSTVKYAAKK MLALKEEKAS DFLDAVSETI 960
961 KTLEGRSRRL KEVLQNSGWE VIQPSAGISM VAKPKAYLNK KVKLKAGDGQ EIVELTDSNM1020
1021 RDVFLSHTGV CLNSGSWTGI PGYCRFSFAL EDSEFDKAIE SIAQFKSVLA N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
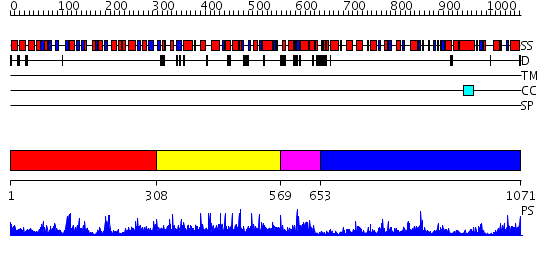
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..307] | 58.0 | Crystal Structure Analysis of E.coli Protein (N5)-Glutamine Methyltransferase (HemK) | |
| 2 | View Details | [308..568] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [569..652] | 4.69897 | Solution structure of Streptmycal repressor TraR | |
| 4 | View Details | [653..1071] | 81.69897 | Aromatic aminoacid aminotransferase, AroAT |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
|||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)