













| General Information: |
|
| Name(s) found: |
VPS11_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 932 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plant-type vacuole membrane
[IDA]
|
| Biological Process: |
vacuole organization
[TAS]
|
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] binding [IEA] transporter activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYQLRKFDFF EEKYGGKIPE DVTGDIQCCS SGRGKVVIGS NDGSVSFLDR GVKFDSGFQA 60
61 HSSSVLFLQH LKQRNFLVTV GEDEQISPQQ SGMCLKVFDL DKVQEEGTSS SAPECIGILR 120
121 IFTNQFPEAK ITSFLVLEEV PPILLIAIGL DNGCIYCVKG DIARERITRF KLQVDGRSAI 180
181 TGLGFRMDGQ ALLLFAVTPE SVNLFSMQAQ PPKLQTLDHI GGSVNTVTMS DRSELIVGRP 240
241 EAVYFYEVDG RGPCWAFEGE KKFMGWFRGY LLCVIDDSKT GNTVFNVYDL RNRLIAYSIV 300
301 VDKVSNMLCE WGNIILIKAD KSLLCITEKD MESKLDMLFK KNLYTVAINL VQSQHADAAA 360
361 TANVMRKYGD HLYGKQDFDE AMLQYINTIG YLEPSFVIQK FLDAQRIYNL TNYLEKLHEK 420
421 GLASKDHTTL LLNCYTKLKD VEKLNTFIRK EDGIGELKFD VETAIRVCRA ANYHEHAMYV 480
481 AKKAGKHEWY LKILLEDLGN YDEALQYVSS LEPSQAGVTI EQYGKILIEH KPKETIDILM 540
541 RLCTEQGIPN GVFLSMLPSP VDFITVFVQH PHSLMHFLER YAEIVQDSPA QAEINNTLLE 600
601 LYLSRDLNFP SISLSENGLD KDLIDHSVAA AVSKADPEKK TNADSKDAME KDCTERQQKG 660
661 LELLKMAWPS DLEQPLYDVD LAVILCEMNS FKDGLLYLYE KMKFYKEVIA CYMQNHDHEG 720
721 LIACCKRLGD SSKGGDPSLW ADLLKYFGEI GEDCTKEVKE VLTYIERDDI LPPIIVLQTL 780
781 AKNPCLTLSV IKDYIARKLE QESKIIEEDR RAVEKYQETT KNMRKEIEDL RTNARIFQLS 840
841 KCTACTFTLD IPAVHFMCMH SFHQRCLGDN EKECPECAPE YRSVMEMKRS LEQNSKDQDL 900
901 FFQQVKGSKD GFSVIAEYFG KGIISKTRDA TS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
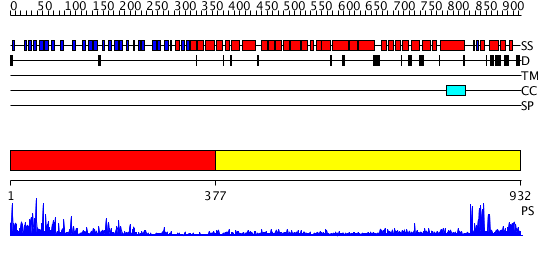
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..376] | 45.522879 | No description for 2gnqA was found. | |
| 2 | View Details | [377..932] | 62.221849 | Clathrin D6 Coat |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)