













| General Information: |
|
| Name(s) found: |
gi|25412332
[NCBI NR]
gi|23506045 [NCBI NR] gi|20465659 [NCBI NR] gi|20197751 [NCBI NR] gi|18175673 [NCBI NR] gi|15224749 [NCBI NR] gi|14194101 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 656 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEFNKEEATR AREIAKRKFL ANDFAGARKF ALKAQFLYPE LDGIAQMVAT FDVHLSAQNI 60
61 IYGDVDHYGV LGLNPEADDE IVRKRYRKLA VMLHPDRNKS VGAEEAFKFL SQAWGVFSDK 120
121 AKRADYDLKR NVGLYKGGGA SSSRPATNGF QKVTKASGNT TKVKSSKRGI KRASDASAAA 180
181 TTSTSAQKTT ADGTFWTVCR TCRTQYEYHS VYLNQNLLCP NCRKPFIAVE TDPPGSGSIR 240
241 KTFHEHQFDS LRHTTDGRKK NVPGRDNNGV YGEYDSFEWG VFTGTKTSAH ATPTGSRKDE 300
301 VVRREYTKRT AGPSSTIPPK RRKVMENAVA GGNIASCLAP KSTGVKEVSE DELKNLLKKK 360
361 AKSVISRNLP ELCTIVAETE TNGRGMETED LNGFNAGSSV NKNAIESCCM NTDKELNSLG 420
421 ALTLDVTAPD FCDFDKDRTE KSVKDNQIWA FYDSHEGLPR SYALIHNVIS VDPFKVRMSW 480
481 LTPVTNGEPS STNWLGFGIP KSCGGFRVRK TLIYRSPYSF SHKVNLVKGN HGEFLIYPRT 540
541 GDVWALYRKW SPDWNYLTGV ETVEYDIVEV VEGYTEEYGV VVVPLVKVAG FKAVFHHHLD 600
601 SKETKRFLRD EISRFSHKIP SYLLTGQEAP GAPRGCRQLD PAATPSQLLQ AIDDYR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
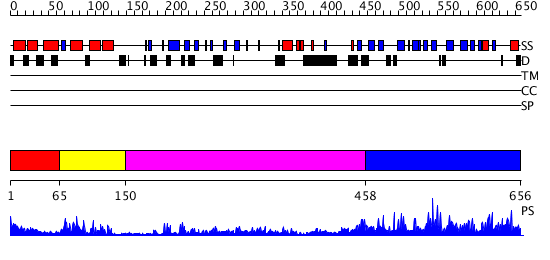
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..64] | 2.186993 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [65..149] | 19.0 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [150..457] | 19.39794 | The crystal structure of Hsp40 Ydj1 | |
| 4 | View Details | [458..656] | 2.091953 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)