













| General Information: |
|
| Name(s) found: |
FAO3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 746 amino acids |
Gene Ontology: |
|
| Cellular Component: |
microsome
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
electron carrier activity
[ISS]
long-chain-alcohol oxidase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKYKVAGKF GLPDITVAEM ESLASFCEAV LPSVQPPPEE LSGEGDNHRN KEALRSFYST 60
61 SGSKTPVLRQ SIELVTKRGT IEAYIATRLI LFLLATRLGT LLICGTECLV SRWPFVEKFS 120
121 ELSLEKRERV LQKQFKNWIL TPIRAAFVYI KVAFLFCFFS RVNPNGENPA WEAIGYRVNP 180
181 DENKPSETHN ERPLEKGIVE TMEETEQTLL ESLAHKGLEA VLDTEHDAIR IKCDVVVVGS 240
241 GSGGGVAASV LAKSGLKVVV LEKGSYFTPS EHRPFEGPGL DKLYENGGIL PSVDGSFMVL 300
301 AGATVGGGSA VNWSACIKTP KSVLQEWSED QNIPLFGTKE YLTAMEVVWK RMGVTEKCEL 360
361 ESFQNQILRK GCENLGFNVE NVPRNSSESH YCGSCGYGCR QGDKKGSDRT WLVDAVGHGA 420
421 VILTGCKAER FILEKNGSNK GGKQMKCLGV MAKSLNGNIA KMLKIEAKVT VSAGGALLTP 480
481 PLMISSGLRN RNIGKNLHLH PVLMAWGYFP DKESSNISFK GNSYEGGIIT SVSKVLSEDS 540
541 EVRAIIETPQ LGPGSFSVLT PWTSGLDMKK RMARYSRTAS LITIVRDRGS GEVKTEGRIN 600
601 YTVDKTDRDN LKAGLRESLR ILIAAGAEEV GTHRSDGQRL ICKGVNENSI QEFLDSVSTE 660
661 EGAKGMTEKW NVYSSAHQMG SCRIGENEKE GAIDLNGESW EAEKLFVCDA SALPSAVGVN 720
721 PMITVMSTAY CISTRIAKSM TTGLSH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
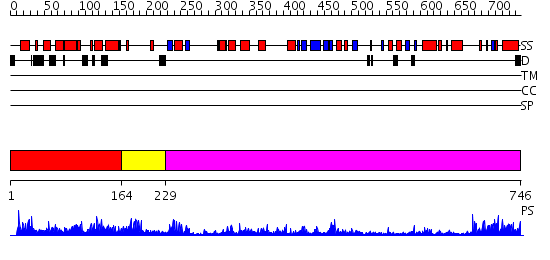
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..163] | 1.277971 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [164..228] | 10.221849 | Dihydropyrimidine dehydrogenase, N-terminal domain; Dihydropyrimidine dehydrogenase, domain 4; Dihydropyrimidine dehydrogenase, domain 3; Dihydropyrimidine dehydrogenase, domain 2; Dihydropyrimidine dehydrogenase, C-terminal domain | |
| 3 | View Details | [229..746] | 59.39794 | Glucose oxidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.43 |
Source: Reynolds et al. (2008)