













| General Information: |
|
| Name(s) found: |
RH27_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 633 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] ATP-dependent helicase activity [IEA][ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANLDMEQHS SENEEIKKKK HKKRARDEAK KLKQPAMEEE PDHEDGDAKE NNALIDEEPK 60
61 KKKKKKNKKR GDTDDGEDEA VAEEEPKKKK KKNKKLQQRG DTNDEEDEVI AEEEEPKKKK 120
121 KKQRKDTEAK SEEEEVEDKE EEKKLEETSI MTNKTFESLS LSDNTYKSIK EMGFARMTQI 180
181 QAKAIPPLMM GEDVLGAART GSGKTLAFLI PAVELLYRVK FTPRNGTGVL VICPTRELAI 240
241 QSYGVAKELL KYHSQTVGKV IGGEKRKTEA EILAKGVNLL VATPGRLLDH LENTNGFIFK 300
301 NLKFLVMDEA DRILEQNFEE DLKKILNLLP KTRQTSLFSA TQSAKVEDLA RVSLTSPVYI 360
361 DVDEGRKEVT NEGLEQGYCV VPSAMRLLFL LTFLKRFQGK KKIMVFFSTC KSTKFHAELF 420
421 RYIKFDCLEI RGGIDQNKRT PTFLQFIKAE TGILLCTNVA ARGLDFPHVD WIVQYDPPDN 480
481 PTDYIHRVGR TARGEGAKGK ALLVLTPQEL KFIQYLKAAK IPVEEHEFEE KKLLDVKPFV 540
541 ENLISENYAL KESAKEAYKT YISGYDSHSM KDVFNVHQLN LTEVATSFGF SDPPKVALKI 600
601 DRGGYRSKRE PVNKFKRGRG GGRPGGKSKF ERY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
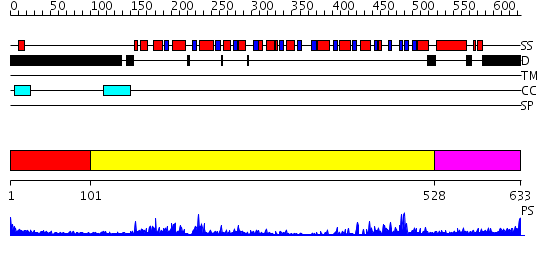
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [101..527] | 82.045757 | No description for 2db3A was found. | |
| 3 | View Details | [528..633] | 26.0 | Nucleotide excision repair enzyme UvrB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)