













| General Information: |
|
| Name(s) found: |
DCE3_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 500 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
glutamate metabolic process
[IEA]
carboxylic acid metabolic process [IEA] |
| Molecular Function: |
calmodulin binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLSKTASKS DDSIHSTFAS RYVRNSISRF EIPKNSIPKE AAYQIINDEL KFDGNPRLNL 60
61 ASFVTTWMEP ECDKLMMESI NKNNVEMDQY PVTTDLQNRC VNMIARLFNA PLGDGEAAIG 120
121 VGTVGSSEAV MLAGLAFKRQ WQNKRKALGL PYDRPNIVTG ANIQVCLEKF ARYFEVELKE 180
181 VKLREGYYVM DPDKAVEMVD ENTICVVAIL GSTLTGEFED VKLLNDLLVE KNKKTGWDTP 240
241 IHVDAASGGF IAPFLYPDLE WDFRLPLVKS INVSGHKYGL VYAGIGWVVW RTKTDLPDEL 300
301 IFHINYLGAD QPTFTLNFSK GSSQVIAQYY QLIRLGFEGY RNVMDNCREN MMVLRQGLEK 360
361 TGRFNIVSKE NGVPLVAFSL KDSSRHNEFE VAEMLRRFGW IVPAYTMPAD AQHVTVLRVV 420
421 IREDFSRTLA ERLVADFEKV LHELDTLPAR VHAKMASGKV NGVKKTPEET QREVTAYWKK 480
481 FVDTKTDKNG VPLVASITNQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
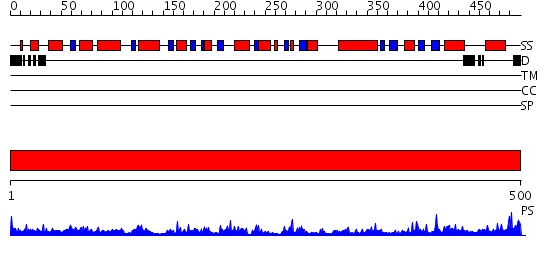
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..500] | 72.0 | Crystal structure of the complex of Escherichia coli GADA with glutarate at 2.05 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)