













| General Information: |
|
| Name(s) found: |
FIMB5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
actin binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSYVGVLVS DPWLQSQFTQ VELRTLKSKF VSNKTQLGRF TVGDLPPVFE KLKAFNGTID 60
61 EDEIKSVLDK SYPNADDEVD FEFFLRAFLS VQARGVEKSG GSKGASSFLK TSTTTVHHAI 120
121 NESEKASYVS HVNNYLRDDP FLKSYLPIDP ATNAFFDLVK DGVLLCKLIN VAVPGTIDER 180
181 AINTKKTLNP WERNENLTLG LNSAKAIGCT VVNIGTQDIA EGRPYLVLGL ISQIIKIQML 240
241 ADLNFKKTPS LFQLVDDTQD AEELMGLAPE KVLLKWMNFH LKKAGYEKQV TNFSSDLKDG 300
301 EAYAYLLNAL APEHSTHVAL ETKDPTERAK KVLEQAEKLD CKRYLSPKDI VDGSANLNLA 360
361 FVAQIFQHRN GLTVDDSKTS FAEMMTDDVE TSREERCFRL WINSLGTATY VNNVFEDLRN 420
421 GWVLLEVLDK VSPGSVNWKH ANKPPIKMPF KKVENCNEVI KIGKELRFSL VNVAGNDIVQ 480
481 GNKKLLLAFL WQLMRYTMLQ LLRNLRSHSQ GKEITDADIL NWANRKVKRG GRTSQADSFR 540
541 DKNLSSGMFF LELLSAVEPR VVNWSLVTNG ETEEDKKLNA TYIISVARKL GCSIFLLPED 600
601 IIEVNQKMML ILAASIMYWS LQQQSDTEST VSEDATDDGD ANSVAGEISN LSIDGASESS 660
661 PTVQDQELLT KADNDEDEVD GENNKDA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
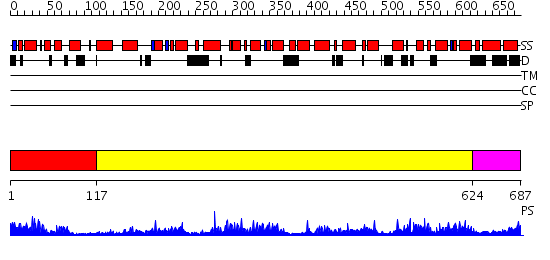
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | 22.69897 | Structure of calmodulin bound to a calcineurin peptide: a new way of making an old binding mode | |
| 2 | View Details | [117..623] | 116.0 | Crystal structure of the actin-crosslinking core of Arabidopsis fimbrin | |
| 3 | View Details | [624..687] | 68.69897 | Cryo-EM Structure of Chicken Gizzard Smooth Muscle alpha-Actinin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)