













| General Information: |
|
| Name(s) found: |
UBP5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 924 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
|
| Biological Process: |
ubiquitin-dependent protein catabolic process
[TAS]
|
| Molecular Function: |
ubiquitin-specific protease activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEVSMGSSS SSTDLSPEEE RVFIRDIAIA AEANSKEGDT FYLITQRWWQ EWIEYVNQDQ 60
61 PCNTNDGSSL SEHCDSPGSS TLKKPSRIDN SDLIYDSSLE DPSNTSEIIE TLQEGRDYVL 120
121 LPQEVWNQLR SWYGGGPTLA RRVISSGLSQ TELAVEVYPL RLQLLLMPKS DHSAIRISKK 180
181 ETIRELHRRA CEIFDLDSEH VRIWDYYGHQ KYSLMNDLDK TLDDANLQMD QDILVEVLDI 240
241 NGTLSSAHIQ SAQENGLVDG DSTSILIEPS KSSLAAAGGF SSSRNAFRTG SVEVSQSFDN 300
301 TYSSTGVTTR GSTAGLTGLL NLGNTCFMNS AIQCLVHTPE FASYFQEDYH QEINWQNPLG 360
361 MVGELALAFG DLLRKLWAPG RTPIAPRPFK AKLARFAPQF SGYNQHDSQE LLAFLLDGLH 420
421 EDLNRVKHKP YINSRDADGR PDEEVADEFW KNHIARNDSI IVDVCQGQYK STLVCPICNK 480
481 VSVTFDPFMY LSLPLQFNTT RAITVTVFSC DKTALPSTIT VNVSKQGRCR DLIQALTNAC 540
541 SLKQSEELKL AEIRNNFIHR LFEDPLIPLS SIKDDDHLAA YKLSKSSENT TLLRLVLRRR 600
601 DQKAGEREST VQLKPCGTPL LSSASCGDAL TKGKIHCLVQ NMLSPFRREE SVGKKGNSDS 660
661 SIPERRSARF NNTEEEDKVG GLKKAKKSNS SDLGASKLSL QLIDEDNKTI NLPDNEAEAM 720
721 KLPSSATVTI YLDWTPELSG MYDITCLESL PEVLKYGPTT KKARSEPLSL YACLEAFLRE 780
781 EPLVPDEMWF CPQCNERRQA SKKLDLWRLP EVLVIHLKRF SYSRSMKHKL ETFVNFPIHD 840
841 LDLTKYVANK NLSQPQLYEL YALTNHYGGM GSGHYTAHIK LLDDSRWYNF DDSHISHINE 900
901 DDVKSGAAYV LFYRRKSDAG GKMT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
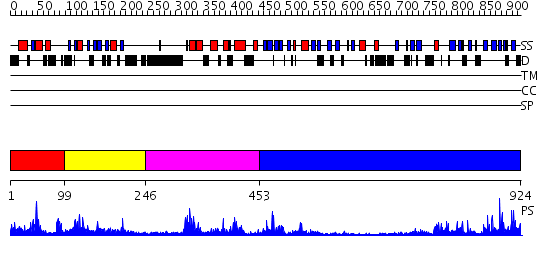
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 30.154902 | Solution structure of the DUSP domain of hUSP15 | |
| 2 | View Details | [99..245] | 5.0 | No description for 2i50A was found. | |
| 3 | View Details | [246..452] | 65.69897 | No description for 2gfoA was found. | |
| 4 | View Details | [453..924] | 47.39794 | Crystal structure of HAUSP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)