













| General Information: |
|
| Name(s) found: |
AKH1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 911 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: |
aspartate family amino acid biosynthetic process
[TAS]
|
| Molecular Function: |
aspartate kinase activity
[IDA]
homoserine dehydrogenase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPVVSLAKVV TSPAVAGDLA VRVPFIYGKR LVSNRVSFGK LRRRSCIGQC VRSELQSPRV 60
61 LGSVTDLALD NSVENGHLPK GDSWAVHKFG GTCVGNSERI KDVAAVVVKD DSERKLVVVS 120
121 AMSKVTDMMY DLIHRAESRD DSYLSALSGV LEKHRATAVD LLDGDELSSF LARLNDDINN 180
181 LKAMLRAIYI AGHATESFSD FVVGHGELWS AQMLAAVVRK SGLDCTWMDA RDVLVVIPTS 240
241 SNQVDPDFVE SEKRLEKWFT QNSAKIIIAT GFIASTPQNI PTTLKRDGSD FSAAIMSALF 300
301 RSHQLTIWTD VDGVYSADPR KVSEAVVLKT LSYQEAWEMS YFGANVLHPR TIIPVMKYDI 360
361 PIVIRNIFNL SAPGTMICRQ IDDEDGFKLD APVKGFATID NLALVNVEGT GMAGVPGTAS 420
421 AIFSAVKEVG ANVIMISQAS SEHSVCFAVP EKEVKAVSEA LNSRFRQALA GGRLSQIEII 480
481 PNCSILAAVG QKMASTPGVS ATFFNALAKA NINIRAIAQG CSEFNITVVV KREDCIRALR 540
541 AVHSRFYLSR TTLAVGIIGP GLIGGTLLDQ IRDQAAVLKE EFKIDLRVIG ITGSSKMLMS 600
601 ESGIDLSRWR ELMKEEGEKA DMEKFTQYVK GNHFIPNSVM VDCTADADIA SCYYDWLLRG 660
661 IHVVTPNKKA NSGPLDQYLK IRDLQRKSYT HYFYEATVGA GLPIISTLRG LLETGDKILR 720
721 IEGIFSGTLS YLFNNFVGTR SFSEVVAEAK QAGFTEPDPR DDLSGTDVAR KVTILARESG 780
781 LKLDLEGLPV QNLVPKPLQA CASAEEFMEK LPQFDEELSK QREEAEAAGE VLRYVGVVDA 840
841 VEKKGTVELK RYKKDHPFAQ LSGADNIIAF TTKRYKEQPL IVRGPGAGAQ VTAGGIFSDI 900
901 LRLAFYLGAP S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
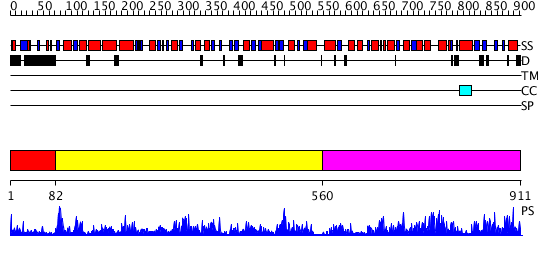
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [82..559] | 114.0 | No description for 3c1mA was found. | |
| 3 | View Details | [560..911] | 67.0 | Homoserine dehydrogenase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)