













| General Information: |
|
| Name(s) found: |
M3KE1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1368 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
plasma membrane [IDA] |
| Biological Process: |
plasma membrane organization
[IGI]
pollen development [IGI] |
| Molecular Function: |
ATP binding
[IEA]
protein tyrosine kinase activity [IEA] protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] binding [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARQMTSSQF HKSKTLDNKY MLGDEIGKGA YGRVYKGLDL ENGDFVAIKQ VSLENIVQED 60
61 LNTIMQEIDL LKNLNHKNIV KYLGSSKTKT HLHIILEYVE NGSLANIIKP NKFGPFPESL 120
121 VAVYIAQVLE GLVYLHEQGV IHRDIKGANI LTTKEGLVKL ADFGVATKLN EADVNTHSVV 180
181 GTPYWMAPEV IEMSGVCAAS DIWSVGCTVI ELLTCVPPYY DLQPMPALFR IVQDDNPPIP 240
241 DSLSPDITDF LRQCFKKDSR QRPDAKTLLS HPWIRNSRRA LQSSLRHSGT IKYMKEATAS 300
301 SEKDDEGSQD AAESLSGENV GISKTDSKSK LPLVGVSSFR SEKDQSTPSD LGEEGTDNSE 360
361 DDIMSDQVPT LSIHEKSSDA KGTPQDVSDF HGKSERGETP ENLVTETSEA RKNTSAIKHV 420
421 GKELSIPVDQ TSHSFGRKGE ERGIRKAVKT PSSVSGNELA RFSDPPGDAS LHDLFHPLDK 480
481 VSEGKPNEAS TSMPTSNVNQ GDSPVADGGK NDLATKLRAT IAQKQMEGET GHSNDGGDLF 540
541 RLMMGVLKDD VIDIDGLVFD EKVPAENLFP LQAVEFSRLV SSLRPDESED AIVSSCQKLV 600
601 AMFRQRPEQK VVFVTQHGFL PLMDLLDIPK SRVICAVLQL INEIIKDNTD FQENACLVGL 660
661 IPVVMSFAGP ERDRSREIRK EAAYFLQQLC QSSPLTLQMF IACRGIPVLV GFLEADYAKY 720
721 REMVHLAIDG MWQVFKLKRS TPRNDFCRIA AKNGILLRLI NTLYSLNEAT RLASISGGLD 780
781 GQAPRVRSGQ LDPNNPIFGQ NETSSLSMID QPDVLKTRHG GGEEPSHAST SNSQRSDVHQ 840
841 PDALHPDGDK PRVSSVAPDA STSGTEDVRQ QHRISLSANR TSTDKLQKLA EGASNGFPVT 900
901 QTEQVRPLLS LLDKEPPSRH YSGQLDYVKH ITGIERHESR LPLLHGSNEK KNNGDLDFLM 960
961 AEFAEVSGRG KENGSLDTTT RYPSKTMTKK VLAIEGVAST SGIASQTASG VLSGSGVLNA1020
1021 RPGSATSSGL LAHMVSTLSA DVAREYLEKV ADLLLEFARA DTTVKSYMCS QSLLSRLFQM1080
1081 FNRVEPPILL KILECTNHLS TDPNCLENLQ RADAIKHLIP NLELKDGHLV YQIHHEVLSA1140
1141 LFNLCKINKR RQEQAAENGI IPHLMLFIMS DSPLKQYALP LLCDMAHASR NSREQLRAHG1200
1201 GLDVYLSLLD DEYWSVIALD SIAVCLAQDN DNRKVEQALL KQDAIQKLVD FFQSCPERHF1260
1261 VHILEPFLKI ITKSYRINKT LAVNGLTPLL ISRLDHQDAI ARLNLLKLIK AVYEHHPRPK1320
1321 QLIVENDLPQ KLQNLIEERR DGQRSGGQVL VKQMATSLLK ALHINTIL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
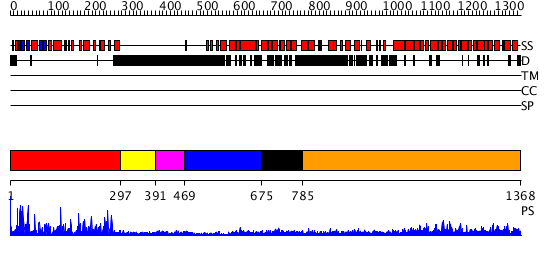
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..296] | 71.39794 | No description for 3comA was found. | |
| 2 | View Details | [297..390] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [391..468] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [469..674] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [675..784] | 5.69897 | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | |
| 6 | View Details | [785..1368] | 48.69897 | No description for 2z6gA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)