













| General Information: |
|
| Name(s) found: |
MCM2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 936 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development
[IMP]
root development [IMP] DNA replication initiation [ISS] regulation of root meristem growth [IMP] DNA unwinding involved in replication [TAS] regulation of cell proliferation [IMP] |
| Molecular Function: |
DNA binding
[IEA][ISS]
ATP binding [IEA][ISS] DNA-dependent ATPase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGENSDNEP SSPASPSSAG FNTDQLPIST SQNSENFSDE EEAAVDTQVI RDEPDEAEDE 60
61 EEEEGEDLFN DTFMNDYRKM DENDQYESNG IDDSVDDERD LGQAMLDRRA ADADLDAREN 120
121 RLANRKLPHL LHDNDSDDWN YRPSKRSRTT VPPRGNGGDP DGNPPSSPGV SQPDISMTDQ 180
181 TDDYQDEDDN DDEAEFEMYR IQGTLREWVM RDEVRRFIAK KFKDFLLTYV KPKNENGDIE 240
241 YVRLINEMVS ANKCSLEIDY KEFIHVHPNI AIWLADAPQP VLEVMEEVSE KVIFDLHPNY 300
301 KNIHTKIYVR VTNLPVNDQI RNIRQIHLNT MIRIGGVVTR RSGVFPQLQQ VKYDCNKCGA 360
361 VLGPFFQNSY SEVKVGSCSE CQSKGPFTVN VEQTIYRNYQ KLTIQESPGT VPAGRLPRHK 420
421 EVILLNDLID CARPGEEIEV TGIYTNNFDL SLNTKNGFPV FATVVEANYV TKKQDLFSAY 480
481 KLTQEDKTQI EELSKDPRIV ERIIKSIAPS IYGHEDIKTA LALAMFGGQE KNIKGKHRLR 540
541 GDINVLLLGD PGTAKSQFLK YVEKTGQRAV YTTGKGASAV GLTAAVHKDP VTREWTLEGG 600
601 ALVLADRGIC LIDEFDKMND QDRVSIHEAM EQQSISISKA GIVTSLQARC SVIAAANPVG 660
661 GRYDSSKSFA QNVELTDPIL SRFDILCVVK DVVDPVTDEM LAEFVVNSHF KSQPKGGKME 720
721 DSDPEDGIQG SSGSTDPEVL PQNLLKKYLT YSKLYVFPKL GELDAKKLET VYANLRRESM 780
781 NGQGVSIATR HLESMIRMSE AHARMHLRQY VTEEDVNMAI RVLLDSFIST QKFGVQRTLR 840
841 ESFKRYITYK KDFNSLLLVL LKELVKNALK FEEIISGSNS GLPTIEVKIE ELQTKAKEYD 900
901 IADLRPFFSS TDFSKAHFEL DHGRGMIKCP KRLITW |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
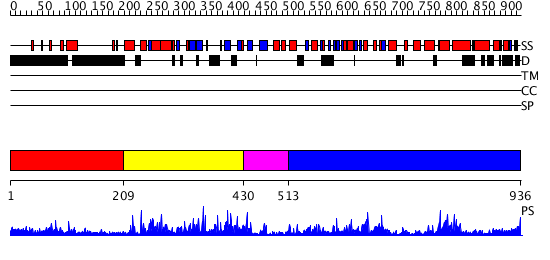
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..208] | 4.118996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [209..429] | 55.39794 | No description for 2vl6A was found. | |
| 3 | View Details | [430..512] | 45.69897 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 4 | View Details | [513..936] | 47.045757 | No description for 2r44A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)