













| General Information: |
|
| Name(s) found: |
NRX1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 578 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
pollen tube growth
[IMP]
pollen tube guidance [IMP] response to cadmium ion [IEP] |
| Molecular Function: |
antioxidant activity
[IEA]
oxidoreductase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAETSKQVNG DDAQDLHSLL SSPARDFLVR NDGEQVKVDS LLGKKIGLYF SAAWCGPCQR 60
61 FTPQLVEVYN ELSSKVGFEI VFVSGDEDEE SFGDYFRKMP WLAVPFTDSE TRDRLDELFK 120
121 VRGIPNLVMV DDHGKLVNEN GVGVIRSYGA DAYPFTPEKM KEIKEDEDRA RRGQTLRSVL 180
181 VTPSRDFVIS PDGNKVPVSE LEGKTIGLLF SVASYRKCTE LTPKLVEFYT KLKENKEDFE 240
241 IVLISLEDDE ESFNQDFKTK PWLALPFNDK SGSKLARHFM LSTLPTLVIL GPDGKTRHSN 300
301 VAEAIDDYGV LAYPFTPEKF QELKELEKAK VEAQTLESLL VSGDLNYVLG KDGAKVLVSD 360
361 LVGKTILMYF SAHWCPPCRA FTPKLVEVYK QIKERNEAFE LIFISSDRDQ ESFDEYYSQM 420
421 PWLALPFGDP RKASLAKTFK VGGIPMLAAL GPTGQTVTKE ARDLVVAHGA DAYPFTEERL 480
481 KEIEAKYDEI AKDWPKKVKH VLHEEHELEL TRVQVYTCDK CEEEGTIWSY HCDECDFDLH 540
541 AKCALNEDTK ENGDEAVKVG GDESKDGWVC EGNVCTKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
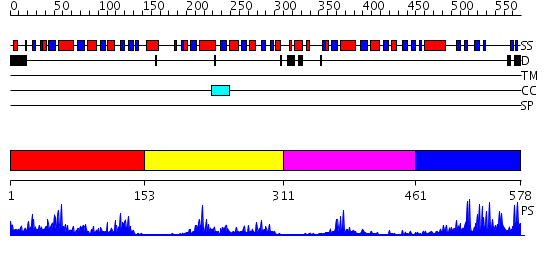
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | 19.30103 | Tryparedoxin I | |
| 2 | View Details | [153..310] | 10.0 | No description for 2h30A was found. | |
| 3 | View Details | [311..460] | 5.86 | No description for 2h30A was found. | |
| 4 | View Details | [461..578] | 14.39794 | Solution Structure of DC1 Domain of PDI-like Hypothetical Protein from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)