













| General Information: |
|
| Name(s) found: |
DNJ10_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 398 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKETEYYDV LGVSPTATES EIKKAYYIKA RQVHPDKNPN DPQAAHNFQV LGEAYQVLSD 60
61 SGQRQAYDAC GKSGISTDAI IDPAAIFAML FGSELFEGYI GQLAMASMAS LDIFTEGDQF 120
121 DTKKIQEKLR IVQKEREDKL AQILKDRLNE YVINKDEFIS NAEAEVARLS NAAYGVDMLN 180
181 TIGYIYVRQA AKELGKKAIY LGVPFIAEWF RNKGHFIKSQ LTAATGAYAL FQLQEEMKRQ 240
241 LNTEGNYTEE ELEEYLQAHK RVMIDSLWKL NVADIEATLC RVCQLVLQDP EAKREELRTR 300
301 ARGLKALGRI FQRAKTASES DPLENSEPQK LNGNGKNHDE DTSTSPKSSE ASHSTSGPQE 360
361 PQSPYVEEFK LGDEQFNYYF PRPAPPPGAG KYSSSGYD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
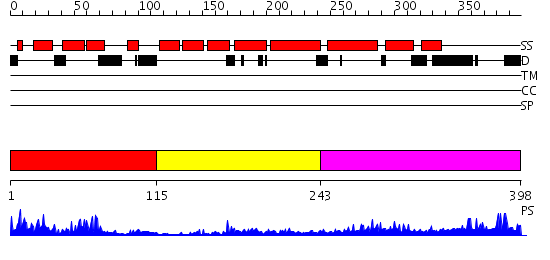
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | 26.69897 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [115..242] | 4.39794 | Solution structure of zinc finger domain from human DnaJ subfamily A menber 3 | |
| 3 | View Details | [243..398] | 2.429989 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)