













| General Information: |
|
| Name(s) found: |
HEM13_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 524 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA][ISS]
cytoplasm [IEA] |
| Biological Process: |
porphyrin biosynthetic process
[TAS]
|
| Molecular Function: |
NADP or NADPH binding
[IEA]
catalytic activity [IEA] glutamyl-tRNA reductase activity [IEA][ISS] binding [IEA] shikimate 5-dehydrogenase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVSNASVVL SPNLETSSSW YHHNPSSSLD LIRIHTLPMN KMTRRGLIQR VRCETSPSSD 60
61 SSPLDKLKNS PAIDRYTRER SSIVVIGLSF HTSPLEMRER LAIPEAEWPL AITQLCALNH 120
121 IEEAAVLSTC NRIEIYVSAL SRYRGVKEVT EWMSKRSGIP VSDICQHRFL LYNKDATQHL 180
181 FQVSAGLESL VIGENQIQSQ VRKAEQVVKQ EGFGRIISTL FEKANKAGKR VRAQTNIASG 240
241 AVSVSSAAVE LALTKLPGSV SSAMMLVIGA GEMGKRIIEH LVAKGCTKMV VMNRSEDKVA 300
301 AIRKEMQSGV EIIYKPLDEI LACAAEANVI FTSTSSETPL FLKEHVEILP PCPADYARLF 360
361 VDISVPRNVG SCVAELDSAR VYNVDDLKEV VAANKEDRAR KSMEALPIIR EETIEFEGWR 420
421 DSLQTFPTIR KLRSKTERIR AECVEKLISK HGNGMDKKTR EAVEKQTRII VNNILDYPMK 480
481 HLRYDGTGSS KLRETLENMQ AVNRIYELDG ELLEEKIREK KDKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
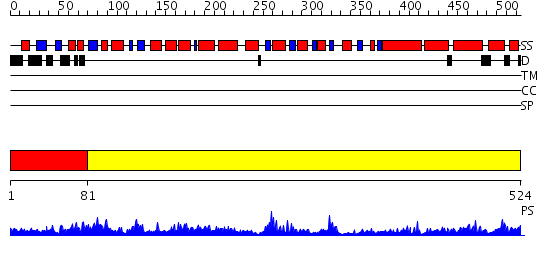
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..524] | 57.522879 | Glutamyl tRNA-reductase dimerization domain; Glutamyl tRNA-reductase middle domain; Glutamyl tRNA-reductase catalytic, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)