













| General Information: |
|
| Name(s) found: |
gi|15240503
[NCBI NR]
gi|10177619 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 689 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
transcription
[IEA]
|
| Molecular Function: |
DNA-directed RNA polymerase activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFDDDDKPK EVTKTRRFAP GRAGKSKPKP KPEPTADKPV QPPPQSQTES VSKTEHDVDA 60
61 KFVGTKVETE VCNGSVKMEI DSKVDKEPEI METELMEEDQ QLPLQEEKEE EEEEDDVVVR 120
121 EIDVFFKPSI DANTQLYVLQ YPLRPSWRPY EMDERCEEVR VNPSTSQVEI DLSMDVHSKN 180
181 YDSNFGLNMT KQTLKTTWKQ PPTLDYAVGV LSGDKLHLNP VHAVAQLRPS MQSLSSDKKK 240
241 KQEESTEESV GTSKKQNKGV QQASTDQKPI NEETWVSLKY HGLQSEYCSR YLNGMMANGN 300
301 SSIDFNMSPG TYINELCRGG SSRNSESKET LKRVLLSLPL KERVQKLLCE GSPLIRYSVL 360
361 KHYAPEFSDE DFLGALQEYG RLVQGLWTPK TRLLKLDGPV EAARDYVLSL FSQNTTIKYS 420
421 EVEATGDKMK PLMERMLTEF AKERHVLKDW KFKEPTDVSF IKSYPEIVKE QDIFWTDKRE 480
481 NLKSRITAQG GKSRADKRRN VVGTSSSVTV KPEVPTTLSD KGGSSKNTIH RVVTQEMPEE 540
541 LKKALPKALK KVFQTHKVCR YETICQGLRD LAVSTSNNPK ADSGMAVNVA LAVDAYQGEL 600
601 EDVINGVATN IHGSYVSISS PDHPEYDSLR EVVISLLTGS PPGTKLMKAE VFAAGRTKLE 660
661 REITNNEYIK VMHEICETNS SGWVLQKAR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
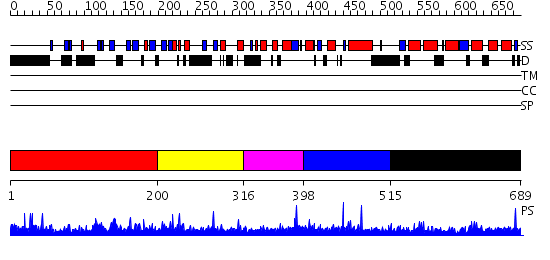
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 0.978 | No description for 2grxC was found. | |
| 2 | View Details | [200..315] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [316..397] | 1.030985 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [398..514] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [515..689] | 1.015979 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)