













| General Information: |
|
| Name(s) found: |
PNSB1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 461 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
NAD(P)H dehydrogenase complex (plastoquinone) [IDA][IMP] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
photosynthetic electron transport in photosystem I
[IMP]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSLPLLPK PISPFFKTPP FSTSKPLVFL NFQTRLTSRS SDVSVNLKKK NNPWLDPFDS 60
61 GEDPDNEYGS LFADGKQDED PRPPDNPDNP YGFLKFPKGY TVELASLPLK IRGDVRRCCC 120
121 VISGGVYENL LFFPTIQLIK DRYPGVQVDI LTTERGKQTY ELNKNVRWAN VYDPDDHWPE 180
181 PAEYTDMIGL LKGRYYDMVL STKLAGLGHA AFLFMTTARD RVSYIYPNVN SAGAGLMLSE 240
241 TFTAENTNLS ELGYSMYTQM EDWLGRPFRS VPRTPLLPLR VSISRKVKEV VAAKYRNAGA 300
301 VTGKFIVIHG IESDSKASMQ SKGDADSLLS LEKWAKIIKG VRGFKPVFVI PHEKERENVE 360
361 DFVGDDTSIV FITTPGQLAA LINDSAGVIA TNTAAIQLAN ARDKPCIGLF SSEEKGKLFV 420
421 PYAEEKSNCV IIASKTGKLA DIDIGTVKNA MQVFEGSLAL V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
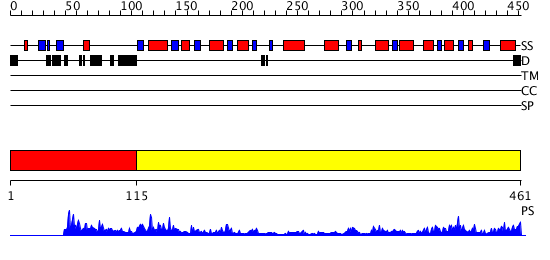
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [115..461] | 54.522879 | ADP-heptose LPS heptosyltransferase II |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)