













| General Information: |
|
| Name(s) found: |
XLG1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 888 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
response to glucose stimulus
[IGI]
response to abscisic acid stimulus [IGI] G-protein coupled receptor protein signaling pathway [ISS] response to fructose stimulus [IGI] response to mannitol stimulus [IGI] response to sucrose stimulus [IGI] |
| Molecular Function: |
guanyl nucleotide binding
[IEA]
signal transducer activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLKEDDCCL FAEEYDGPPL SYNIPCAVPI NVEKIPVAAV VSPVCISDNM SFPVIQPILS 60
61 VESKKFLIDS VSPTSVIANC GSNQLELVSD SITVSPTSVI EHTEEEEEEE GGDGEDCELS 120
121 SSGELLLRSC SVKESLDLNE SSSNPLVPDW ESNESVLSMD YPSSRVTGDC VSETNGDGKK 180
181 QPVVTFLGIA SDDGFEEEES CSNLRRVRVV PVKKQPQTKG KKGSCYRCFK GSRFTEKEVC 240
241 LVCDAKYCNS CVLRAMGSMP EGRKCVTCIG FPIDESKRGS LGKCSRMLKR LLNDLEVKQI 300
301 MKTERFCEAN QLPAEYVYVN GQPLYPEELV TLQTCSNPPK KLKPGDYWYD KVSGLWGKEG 360
361 EKPYQIISPH LNVGGPISPE ASNGNTQVFI NGREITKVEL RMLQLAGVQC AGNPHFWVNE 420
421 DGSYQEEGQK NTKGYIWGKA GTKLLCAVLS LPVPSKSTAN ASGEQLYSAN SRSILDHLEH 480
481 RTLQKILLVG NSGSGTSTIF KQAKILYKDV PFLEDERENI KVIIQTNVYG YLGMLLEGRE 540
541 RFEEEALALR NTKQCVLENI PADEGDAKSN DKTVTMYSIG PRLKAFSDWL LKTMAAGNLG 600
601 VIFPAASREY APLVEELWRD AAIQATYKRR SELGLLPSVA SYFLERAIDV LTPDYEPSDL 660
661 DILYAEGVTS SSGLACLDFS FPQTASEENL DPSDHHDSLL RYQLIRVPSR GLGENCKWID 720
721 MFEDVGMVVF VVSMSDYDQV SEDGTNKMLL TKKLFESIIT HPIFENMDFL LILNKYDLLE 780
781 EKVERVPLAR CEWFQDFNPV VSRHRGSNNG NPTLGQLAFH FMAVKFKRFY SSLTGKKLFV 840
841 SSSKSLDPNS VDSSLKLAME ILKWSEERTN ICMSEYSMYS TEPSSFSN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
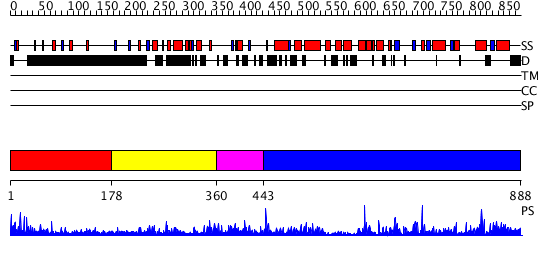
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..177] | 1.02599 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [178..359] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [360..442] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [443..888] | 103.0 | No description for 2hlbA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)