













| General Information: |
|
| Name(s) found: |
ATPD_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 234 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
stromule [IDA] membrane [IDA] chloroplast envelope [IDA] plastoglobule [IDA] thylakoid [IDA] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
response to cold
[IEP]
photosynthetic electron transport in photosystem II [IMP] photosynthesis [IMP] photosynthetic electron transport in photosystem I [IMP] defense response to bacterium [IEP] |
| Molecular Function: |
proton-transporting ATPase activity, rotational mechanism
[IEA]
hydrogen ion transporting ATP synthase activity, rotational mechanism [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASLQQTLFS LQSKLPPSSF QIARSLPLRK TFPIRINNGG NAAGARMSAT AASSYAMALA 60
61 DVAKRNDTME LTVTDIEKLE QVFSDPQVLN FFANPTITVE KKRQVIDDIV KSSSLQSHTS 120
121 NFLNVLVDAN RINIVTEIVK EFELVYNKLT DTQLAEVRSV VKLEAPQLAQ IAKQVQKLTG 180
181 AKNVRVKTVI DASLVAGFTI RYGESGSKLI DMSVKKQLED IASQLELGEI QLAT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
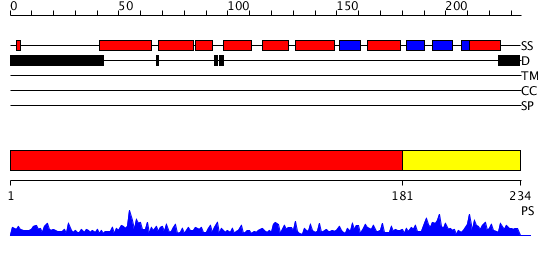
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..180] | 25.69897 | N-TERMINAL DOMAIN OF THE DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE FROM ESCHERICHIA COLI, NMR, MINIMIZED AVERAGE STRUCTURE | |
| 2 | View Details | [181..234] | 1.328994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)