













| General Information: |
|
| Name(s) found: |
UBC23_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1102 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
regulation of protein metabolic process
[IEA]
post-translational protein modification [IEA] |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS]
small conjugating protein ligase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEHEQDDPGT STNVGVDSSV DDSMASLSIC DSEHPNIYRQ DIVKNKKTGS VGVVSEVAGD 60
61 SDSDSDISDE EEDDDDDEDN DDDDEDVEEG KKASEENVVN GDGEKKADGN YKCGALEGDQ 120
121 IRVLWMDNTE PVQDINDVTV IDRGFLHGDY VASASEPTGQ VGVVVDVNIS VDLLAPDGSI 180
181 HKDISTKNLK RVRDFAVGDY VVHGPWLGRI DDVLDNVTVL FDDGSMCKVL RVEPLRLKPI 240
241 PKNNLEEDAN FPYYPGQRVK ASSSSVFKNS RWLSGLWKPN RLEGTVTKVT AGSIFVYWIA 300
301 SAGFGPDSSV SPPEEQNPSN LTLLSCFTHA NWQVGDWCLL PSLNQSATIP LHKHVSKLRL 360
361 YDSQADRQQK IGRDLEDVQD EVSGKVEPAG ITAEALPKVT SDDPPQRNPS VSKEPVHEPW 420
421 PLHRKKIRKL VIRKDKKVKK KEESFEQALL VVNSRTRVDV SWQDGTIECR REAITLIPIE 480
481 TPGDHEFVSE QYVVEKTSDD GDNTTEPRRA GVVKNVNAKD RTASVRWLNP LRRAEEPREF 540
541 EKEEIVSVYE LEGHPDYDYC YGDVVVRLSP IAVALPASSP GNSFEEATQQ DNGYQDSESH 600
601 QEAKILVDKE ENEPSTDLSK LSWVGNITGL KDGDIEVTWA DGTISTVGPH AVYVVGRDDD 660
661 DESVAGESET SDAASWETLN DDDRGAPEIP EEDLGRSSSI EGNSDADIYA ENDSGRNGAL 720
721 ALPLAAIEFV TRLASGIFSR ARKSVDSSSS DYTVENVYKQ AESTNPSDET DSLDDPSPSK 780
781 VNVTDNCESK GTQANAKNIL SGETSTFLED EDKPVPSEGD SCSFRRFDIS QDPLDHHFLG 840
841 VDGQKTKERQ WFKKVDQDWK ILQNNLPDGI FVRAYEDRMD LLRAVIVGAF GTPYQDGLFF 900
901 FDFHLPSDYP SVPPSAYYHS GGWRLNPNLY EEGKVCLSLL NTWTGRGNEV WDPKSSSILQ 960
961 VLVSLQGLVL NSKPYFNEAG YDKQVGTAEG EKNSLGYNEN TFLLNCKTMM YLMRKPPKDF1020
1021 EELIKDHFRK RGYYILKACD AYMKGYLIGS LTKDASVIDE RSSANSTSVG FKLMLAKIAP1080
1081 KLFSALSEVG ADCNEFQHLQ QQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
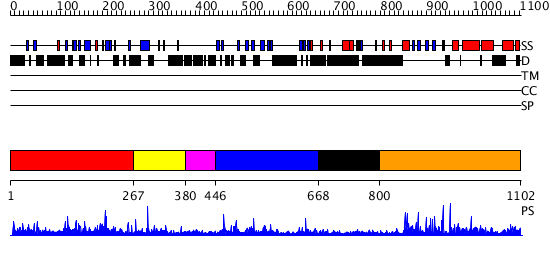
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..266] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [267..379] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [380..445] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [446..667] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [668..799] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [800..1102] | 45.522879 | No description for 3cegA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)