













| General Information: |
|
| Name(s) found: |
TPS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 942 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development
[IMP]
cell division [IMP] embryonic development ending in seed dormancy [IMP] sugar mediated signaling pathway [IMP] trehalose metabolic process [TAS] plant-type cell wall biogenesis [IMP] |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS]
alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity [IGI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPGNKYNCSS SHIPLSRTER LLRDRELREK RKSNRARNPN DVAGSSENSE NDLRLEGDSS 60
61 RQYVEQYLEG AAAAMAHDDA CERQEVRPYN RQRLLVVANR LPVSAVRRGE DSWSLEISAG 120
121 GLVSALLGVK EFEARWIGWA GVNVPDEVGQ KALSKALAEK RCIPVFLDEE IVHQYYNGYC 180
181 NNILWPLFHY LGLPQEDRLA TTRSFQSQFA AYKKANQMFA DVVNEHYEEG DVVWCHDYHL 240
241 MFLPKCLKEY NSKMKVGWFL HTPFPSSEIH RTLPSRSELL RSVLAADLVG FHTYDYARHF 300
301 VSACTRILGL EGTPEGVEDQ GRLTRVAAFP IGIDSDRFIR ALEVPEVIQH MKELKERFAG 360
361 RKVMLGVDRL DMIKGIPQKI LAFEKFLEEN ANWRDKVVLL QIAVPTRTDV PEYQKLTSQV 420
421 HEIVGRINGR FGTLTAVPIH HLDRSLDFHA LCALYAVTDV ALVTSLRDGM NLVSYEFVAC 480
481 QEAKKGVLIL SEFAGAAQSL GAGAILVNPW NITEVAASIG QALNMTAEER EKRHRHNFHH 540
541 VKTHTAQEWA ETFVSELNDT VIEAQLRISK VPPELPQHDA IQRYSKSNNR LLILGFNATL 600
601 TEPVDNQGRR GDQIKEMDLN LHPELKGPLK ALCSDPSTTI VVLSGSSRSV LDKNFGEYDM 660
661 WLAAENGMFL RLTNGEWMTT MPEHLNMEWV DSVKHVFKYF TERTPRSHFE TRDTSLIWNY 720
721 KYADIEFGRL QARDLLQHLW TGPISNASVD VVQGSRSVEV RAVGVTKGAA IDRILGEIVH 780
781 SKSMTTPIDY VLCIGHFLGK DEDVYTFFEP ELPSDMPAIA RSRPSSDSGA KSSSGDRRPP 840
841 SKSTHNNNKS GSKSSSSSNS NNNNKSSQRS LQSERKSGSN HSLGNSRRPS PEKISWNVLD 900
901 LKGENYFSCA VGRTRTNARY LLGSPDDVVC FLEKLADTTS SP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
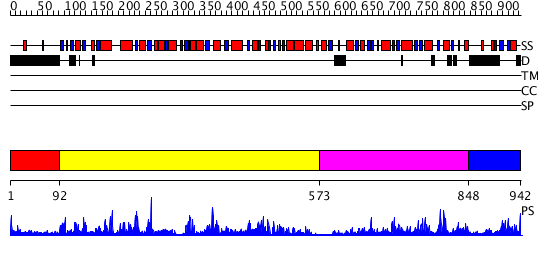
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [92..572] | 79.045757 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 3 | View Details | [573..847] | 26.154902 | Crystal structure of trehalose-6-phosphate phosphatase related protein | |
| 4 | View Details | [848..942] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)