













| General Information: |
|
| Name(s) found: |
MS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 672 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
pollen wall assembly
[IMP]
microgametogenesis [IMP] regulation of transcription, DNA-dependent [ISS] tapetal layer morphogenesis [IMP] pollen germination [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANLIRTDHQ QHIPKKRKRG ESRVFRLKTF GESGHPAEMN ELSFRDNLAK LLEFGHFESS 60
61 GLMGSWSFQL EIQRNPNPLY VLLFVVEEPI EASLNLRCNH CQYVGWGNQM ICNKKYHFVI 120
121 PSKETMAAFL KLEGGGYAFP EKESFSHLVE LQGHVLHGFF HSNGFGHLLS LNGIETGSDL 180
181 TGHQVMDLWD RLCTGLKARK IGLNDASHKK GMELRLLHGV AKGEPWFGRW GYRFGSGTYG 240
241 VTQKIYEKAL ESVRNIPLCL LNHHLTSLNR ETPILLSKYQ SLSTEPLITL SDLFRFMLHL 300
301 HSRLPRDNYM SNSRNQIISI DSTNCRWSQK RIQMAIKVVI ESLKRVEYRW ISRQEVRDAA 360
361 RNYIGDTGLL DFVLKSLGNQ VVGNYLVRRS LNPVKKVLEY SLEDISNLLP SSNNELITLQ 420
421 NQNSMGKMAT NGHNKITRGQ VMKDMFYFYK HILMDYKGVL GPIGGILNQI GMASRAILDA 480
481 KYFIKEYHYI RDTSAKTLHL DRGEELGIFC TIAWKCHHHN NEIKVPPQEC IVVKKDATLS 540
541 EVYGEAERVF RDIYWELRDV VVESVVGGQI EITRVDEMAL NGNKGLVLEG NVGMMMNIEV 600
601 TKCYEDDDKK KDKRIECECG ATEEDGERMV CCDICEVWQH TRCVGVQHNE EVPRIFLCQS 660
661 CDQHLIPLSF LP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
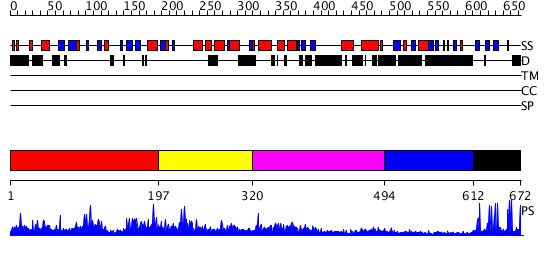
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [197..319] | 2.020965 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [320..493] | 3.414979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [494..611] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [612..672] | 12.69897 | Solution structure of PHD domain in PHD finger family protein |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)