













| General Information: |
|
| Name(s) found: |
SPP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 422 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
sucrose biosynthetic process
[ISS]
response to cadmium ion [IEP] |
| Molecular Function: |
magnesium ion binding
[IEA]
catalytic activity [IEA] sucrose-phosphatase activity [IEA][ISS] phosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERLTSPPRL MIVSDLDHTM VDHHDPENLS LLRFNSLWEH AYRHDSLLVF STGRSPTLYK 60
61 ELRKEKPLLT PDITIMSVGT EITYGNSMVP DHGWVEALNN KWDLGIVKQE ASNFPELKLQ 120
121 AETEQRPHKV SFYVEKSKAQ EVTKELSQRF LKRGLDVKII YSGGMDLDIL PQGAGKGQAL 180
181 AYLLKKLKTE GKLPVNTLAC GDSGNDAELF SIPDVYGVMV SNAQEELLKW HAENAKDNPK 240
241 VIHAKERCAG GIIQAIGHFK LGPNLSPRDV SDFLEIKVEN VNPGHEVVKF FLFYERWRRG 300
301 EVENSEAYTA SLKASVHPGG VFVHPSGTEK SLRDTIDELR KYHGDKQGKK FRVWADQVLA 360
361 TDTTPGTWIV KLDKWEQDGD ERRCCTTTVK FTSKEGEGLV WEHVQQTWSK ETMVKDDSSW 420
421 II |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
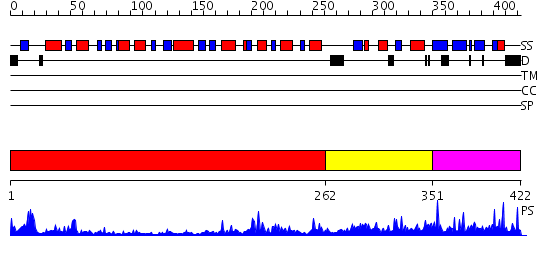
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..261] | 33.30103 | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution | |
| 2 | View Details | [262..350] | 7.39794 | Crystal Structure of the peptidylprolyl isomerase domain of Human PPWD1 | |
| 3 | View Details | [351..422] | 2.67 | Structural Genomics, 1.5A crystal structure of a hypothetical protein PA1314 from Pseudomonas aeruginosa |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)