













| General Information: |
|
| Name(s) found: |
SERA3_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 588 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] |
| Biological Process: |
metabolic process
[IEA]
L-serine biosynthetic process [IEA] |
| Molecular Function: |
oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
[IEA]
cofactor binding [IEA] catalytic activity [IEA] NAD or NADH binding [IEA] phosphoglycerate dehydrogenase activity [IEA] amino acid binding [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATSLNLSSI FSSSSRLVTT PSSVFPIRQR RRIILVTSSS SGGGGKPTIL VTEKLGQAGI 60
61 DLLKKYANVD CSYDLSLEEL CTKISLCDAL IVRSGTKVGR DVFESSRGRL KVVGRAGVGI 120
121 DNVDLAAATE YGCLVVNAPT ANTVAAAEHG IALLTAMARN IAQADASIKA GKWTRNKYVG 180
181 VSLVGKTLAV LGFGKVGSEV ARRARGLGMH VITHDPYAPA DRARAIGVEL VSFEVAISTA 240
241 DFISLHLPLT AATSKMMNDV TFAMMKKGVR IVNVARGGVI DEEALLRALD SGIVAQAALD 300
301 VFTVEPPVKD NKLVLHESVT ATPHLGASTM EAQEGVSIEV AEAVIGALRG ELAATAVNAP 360
361 MVPLEVLREL KPYVVLAEKL GRLAVQLVTG GSGVNAVKVT YASSRAPDDL DTRLLRAMVI 420
421 KGIIEPISSV FINLVNSDYI AKQRGVKISE ERMVLDGSPE NPIEYITVRI ANVESRFASA 480
481 LSESGEIKVE GRVKQGVPSL TKVGLFGVDV SLEGSVILCR QVDQPGMIGK VASILGDENV 540
541 NVSFMSVGRI APGKQAVMAI GVDEQPSKET LKKIGDIPAI EEFVFLKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
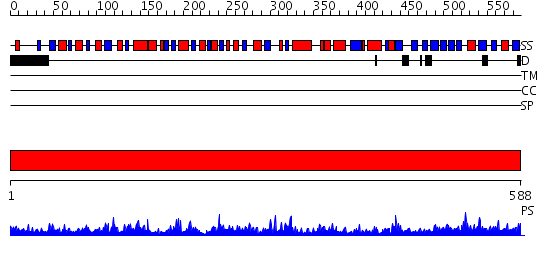
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..588] | 116.0 | Crystal Structure of D-3-Phosphoglycerate dehydrogenase From Mycobacterium tuberculosis |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.49 |
Source: Reynolds et al. (2008)