













| General Information: |
|
| Name(s) found: |
RH33_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 845 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] ATP-dependent helicase activity [IEA][ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMAMRLPAI SRAVTEVASS PVGLRRLFCS NASRFSFLSP PARRQAEPST NLFHSGLSKR 60
61 ITSERSLWNR IFSRNMGGGP RTFPGGLNKW QWKRMHEKKA REKENKLLDQ EKQLYEARIR 120
121 TEIRAKMWGH PDSGEKTAKL KQSHGPMSPK EHIKTLADRF MKAGADDLWN DNDGPVKKFD 180
181 QGSRSCSDSI DSTPIDVRRL VSATCDSMGK HRVLDSSRRG FSSMSRFKRN ESSCDEGDDV 240
241 DAKKLDTLSP FSPKFSGTKE KVKSSTSVVG VIRNKGLFGR RKFRKNDSST EEDSDEEGNE 300
301 GKMIGWMDLR KTGSSASLGN HDIKLTKRVN RNVTDEELYP PLDINRVRED LSKKQSVDNV 360
361 MEEKQEPHDS IYSAKRFDES CISPLTLKAL SASGIVKMTR VQDATLSECL DGKDALVKAK 420
421 TGTGKSMAFL LPAIETVLKA MNSGKGVHKV APIFVLILCP TRELASQIAA EGKALLKNHD 480
481 GIGVQTLIGG TRFRLDQQRL ESEPCQILIA TPGRLLDHIE NKSGLTSRLM ALKLFIVDEA 540
541 DLLLDLGFKR DVEKIIDCLP RQRQSLLFSA TIPKEVRRVS QLVLKRDHSY IDTIGLGCVE 600
601 THDKVKQSCI VAPHESHFHL VPHLLKEHIN NMPDYKIIVF CSTGMVTSLM YTLLREMKLN 660
661 VREIHARKPQ LHRTCVSDEF KESNRLILVT SDVSARGMNY PDVTLVIQVG IPSDREQYIH 720
721 RLGRTGREGK GGKGLLLIAP WERYFLDELK DLPLEPIPAP DLDSRVKHQV DQSMAKIDTS 780
781 IKEAAYHAWL GYYNSVRETG RDKTTLAELA NRFCHSIGLE KPPALFRRTA VKMGLKGISG 840
841 IPIRK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
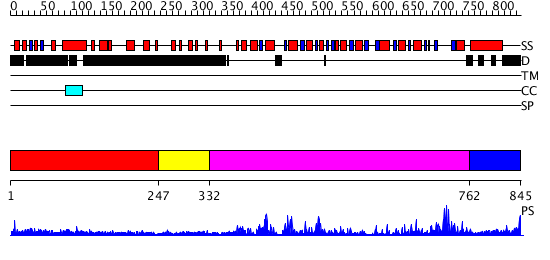
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..246] | 1.317992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [247..331] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [332..761] | 88.221849 | No description for 2db3A was found. | |
| 4 | View Details | [762..845] | 21.69897 | Nucleotide excision repair enzyme UvrB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)