













| General Information: |
|
| Name(s) found: |
DCE4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 493 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] |
| Biological Process: |
glutamate metabolic process
[IEA]
carboxylic acid metabolic process [IEA] |
| Molecular Function: |
calmodulin binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLSKTVSES DVSIHSTFAS RYVRNSLPRF EMPENSIPKE AAYQIINDEL MLDGNPRLNL 60
61 ASFVTTWMEP ECDKLMMESI NKNYVDMDEY PVTTELQNRC VNMIARLFNA PLGDGEAAVG 120
121 VGTVGSSEAI MLAGLAFKRQ WQNKRKAQGL PYDKPNIVTG ANVQVCWEKF ARYFEVELKE 180
181 VNLREDYYVM DPVKAVEMVD ENTICVAAIL GSTLTGEFED VKLLNDLLVE KNKQTGWDTP 240
241 IHVDAASGGF IAPFLYPELE WDFRLPLVKS INVSGHKYGL VYAGIGWVVW RTKTDLPDEL 300
301 IFHINYLGAD QPTFTLNFSK GSSQVIAQYY QLIRLGFEGY RNVMDNCREN MMVLRQGLEK 360
361 TGRFKIVSKE NGVPLVAFSL KDSSRHNEFE VAHTLRRFGW IVPAYTMPAD AQHVTVLRVV 420
421 IREDFSRTLA ERLVADFEKV LHELDTLPAR VHAKMANGKV NGVKKTPEET QREVTAYWKK 480
481 LLETKKTNKN TIC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
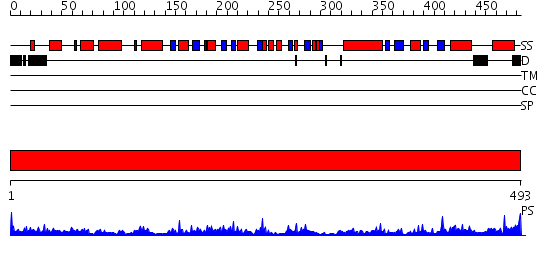
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..493] | 63.69897 | Crystal structure of the complex of Escherichia coli GADA with glutarate at 2.05 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)