













| General Information: |
|
| Name(s) found: |
XLG3_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 848 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
root development
[IGI]
response to glucose stimulus [IGI] response to abscisic acid stimulus [IGI] G-protein coupled receptor protein signaling pathway [ISS] response to fructose stimulus [IGI] response to mannitol stimulus [IGI] gravitropism [IGI] thigmotropism [IGI] response to ethylene stimulus [IMP] response to sucrose stimulus [IGI] |
| Molecular Function: |
guanyl nucleotide binding
[IEA]
signal transducer activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKKDEGESW KEMVRKMLPP GAPLPEDPSE FDYSIALEYT GPPPVHDIPR VSPVDVNPRV 60
61 NNPIPLPVSR IAGGVTSSSG GSPASSESVV SVLHNNPESS SGSASVSPVS GHRQNGNQVR 120
121 RPVVKFKPVD DHDRIEGREA AEEEDNNVEA ETERERKVHE CTASTKRRKK KKKSECYRCG 180
181 KAKWENKETC IVCDEKYCGN CVLRAMGSMP EGRKCVSCIG QAIDESKRSK LGKHSRVLSR 240
241 LLSPLEVKQI MKAEKECTAN QLRPEQLIVN GYPLKPEEMA DLLNCLLPPQ KLKPGRYWYD 300
301 KESGLWGKEG EKPDRVISSN LNFTGKLSPD ASNGNTEVYI NGREITKLEL RILKLANVQC 360
361 PRDTHFWVYD DGRYEEEGQN NIRGNIWEKA STRFMCALFS LPVPQGQPRG TVQPSSNYAT 420
421 VPNYIEHKKI QKLLLLGIEG SGTSTIFKQA KFLYGNKFSV EELQDIKLMV QSNMYRYLSI 480
481 LLDGRERFEE EALSHTRGLN AVEGDSGGEE ANDEGTVTTP QSVYTLNPRL KHFSDWLLDI 540
541 IATGDLDAFF PAATREYAPL VEEVWKDPAI QATYRRKDEL HFLPDVAEYF LSRAMEVSSN 600
601 EYEPSERDIV YAEGVTQGNG LAFMEFSLSD HSPMSESYPE NPDALSSPQP KYQLIRVNAK 660
661 GMNDSCKWVE MFEDVRAVIF CISLSDYDQI NITPESSGTV QYQNKMIQSK ELFESMVKHP 720
721 CFKDTPFILI LNKYDQFEEK LNRAPLTSCD WFSDFCPVRT NNNVQSLAYQ AYFYVAMKFK 780
781 LLYFSITGQK LFVWQARARD RANVDEGFKY VREVLKWDEE KEESYLNGGG EDSFYSTDMS 840
841 SSPYRPEE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
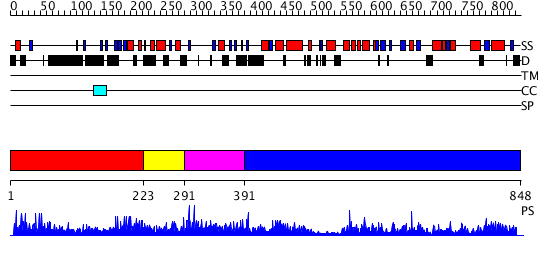
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..222] | 2.4 | Hrs | |
| 2 | View Details | [223..290] | 2.54 | Crystal Structure of a FYVE-type domain from caspase regulator CARP2 | |
| 3 | View Details | [291..390] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [391..848] | 127.0 | Structure of the K180P mutant of Gi alpha subunit bound to AlF4 and GDP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)