













| General Information: |
|
| Name(s) found: |
gi|7487029
[NCBI NR]
gi|7267497 [NCBI NR] gi|3377810 [NCBI NR] gi|15236525 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 587 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
cell redox homeostasis
[IEA]
|
| Molecular Function: |
protein disulfide oxidoreductase activity
[IEA]
electron carrier activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESSPKQQNT EEERKSAEII AKEVSPKQQN VEEERKSAEI IAKEVSPKHN VEKKEEEFTR 60
61 KPVVEIEEEE EEMESIDIHE EEEGDNNVSL DEIMSVDSSD DDDDSESSAE ITKACEETVV 120
121 DERSGISQES NSSTSSAWTE KAAAIKNFVR AKSEVAVHTM IRRLSGKLSI DNAAHGTKDD 180
181 EVESPKTEGK SLWNPLSYLK MMQNDEDLVY REEETVFEPV VMKGRIILYT RLGCEECRGC 240
241 RLFLHEKRLR YVEINIDIYP TRKVELEKIS GGDVVPMVFF NEKLVGSYKE LKVLEESGEL 300
301 EEKIKHLIEE TPPREAPLPP FSGEDDASSK GPVDELALIV LKMKPCVVKD RFYKMRRFKN 360
361 CFLGSEAVDF LSADQRLERD GPRPIVEIAS RLRLVYRAIL EAYTSPDGKH VDYRSIHGSE 420
421 EFARYLRIIQ ELHRVELEDM QREEKLAFFI NLYNMMAIHS ILVWGHPAGT FDRTKMFMDF 480
481 KYVIGGYTYS LSAIQNGILR GNQRPMFNPM KPFGVKDKRS KVALPYAEPL THFTLVCGTR 540
541 SGPPLRCFTP GEIDKELMEA ARDFLRCGGL RVDLNAKVAE ISKIFDW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
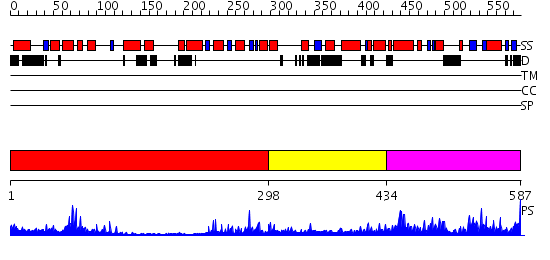
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..297] | 15.69897 | C-terminal, Grx domain of Hybrid-Prx5; N-terminal, Prx domain of Hybrid-Prx5 | |
| 2 | View Details | [298..433] | 1.45 | Solution structure of the DEP domain of human pleckstrin | |
| 3 | View Details | [434..587] | 76.468521 | No description for PF04784.5 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)