













| General Information: |
|
| Name(s) found: |
STIP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 849 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA][ISS]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
nucleic acid binding
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEYQDMERF SMDNDYEGGR WEGDEFVYQK RKEKRKQTKN DATYGIFAES DSDSDDSGGG 60
61 GSRRKRRKDR DSGRKADLTK PVNFVSTGTV MPNQEIDKDS REHNDEKDRD KIEDNDMIDE 120
121 DVEVRGGLGI GSSGLGLGFN ANGFDDEDNL LPGALGKKIA DRAKMRGKAK VEKRGQEGGG 180
181 AKGGKKNTLG SDIGQFEKST KGIGMKLLEK MGYKGGGLGK NQQGIVAPIE AQLRPKNMGM 240
241 GYNDFKEAKL PDLKKVEEKK IIGVSVSENE QSHGDRGGKN LWKKKKVRKA VYVTAEELLE 300
301 KKQEAGFGGG QTIIDMRGPQ VRVVTNLENL DAEEKAKEAD VPMPELQHNL RLIVDLVEHE 360
361 IQKIDRDLRN ERESALSLQQ EKEMLINEEE KQKRHLENME YIADEISRIE LENTSGNLTL 420
421 DSLAIRFEDL QTSYPDDYKL CSLSTIACSL ALPLFIRMFQ GWDPLSDAVH GLKAISSWRK 480
481 LLEVEEDHNI WVVSTPYSQL VSEVVLPAVR IAGINTWEPR DPEPMLRFLE TWETLLPSSV 540
541 LQTILDTVVL PKLSTAVEYW DPRRELVAIH VWVHPWLPIL GQKLEFLYQI IQMKLSNVLD 600
601 AWHPSDSSAY TILSPWKTVF DTTSWEQLMR RYIVPKLQLA LQEFQVNPAN QNLERFDWVM 660
661 KWASAVPIHL MADMMERFFF PKWLDVLYHW LRAKPRFEEI QGWYYGWKEL FPQELTANER 720
721 IRIQLKRGLD MLMEAVEGVE VSQPRAKANE RTQSVPAQAQ AQAKAQMDST EVLSLKEVLE 780
781 VFAQEQELLF KPKPNRMHNG LQIYGFGNVS VIIDSVNQKL LAQKDGGWFL VTPDDLLRMH 840
841 NNTTVSAKR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
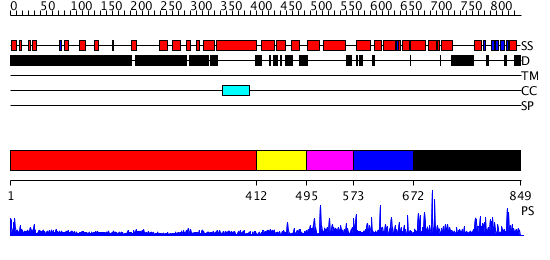
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..411] | 13.221849 | Heavy meromyosin subfragment | |
| 2 | View Details | [412..494] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [495..572] | 1.312994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [573..671] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [672..849] | 2.066957 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)