













| General Information: |
|
| Name(s) found: |
MMSA_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 607 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
response to oxidative stress
[IDA]
|
| Molecular Function: |
3-chloroallyl aldehyde dehydrogenase activity
[ISS]
copper ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVRVKQKNLE SYRSNGTYPP TWRNPTTSFA PDQHRVSIHS SLKSKTKRRR LYKEADDNTK 60
61 LRSSSSTTTT TTTMLLRISG NNLRPLRPQF LALRSSWLST SPEQSTQPQM PPRVPNLIGG 120
121 SFVESQSSSF IDVINPATQE VVSKVPLTTN EEFKAAVSAA KQAFPLWRNT PITTRQRVML 180
181 KFQELIRKNM DKLAMNITTE QGKTLKDSHG DIFRGLEVVE HACGMATLQM GEYLPNVSNG 240
241 VDTYSIREPL GVCAGICPFN FPAMIPLWMF PVAVTCGNTF ILKPSEKDPG ASVILAELAM 300
301 EAGLPDGVLN IVHGTNDTVN AICDDEDIRA VSFVGSNTAG MHIYARAAAK GKRIQSNMGA 360
361 KNHGLVLPDA NIDATLNALL AAGFGAAGQR CMALSTVVFV GDAKSWEDKL VERAKALKVT 420
421 CGSEPDADLG PVISKQAKER ICRLIQSGVD DGAKLLLDGR DIVVPGYEKG NFIGPTILSG 480
481 VTPDMECYKE EIFGPVLVCM QANSFDEAIS IINKNKYGNG AAIFTSSGAA ARKFQMDIEA 540
541 GQIGINVPIP VPLPFFSFTG NKASFAGDLN FYGKAGVDFF TQIKTVTQQW KDIPTSVSLA 600
601 MPTSQKQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
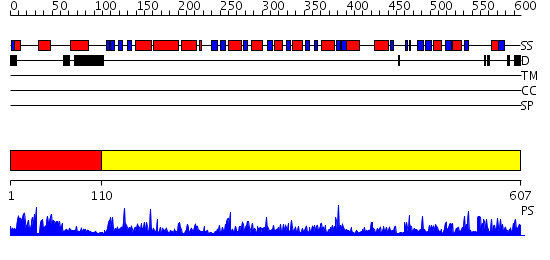
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 37.30103 | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-Tetrahydro-2-furoic acid | |
| 2 | View Details | [110..607] | 145.0 | Crystal structure of methylmalonate semialdehyde dehydrogenase from Bacillus subtilis |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)