













| General Information: |
|
| Name(s) found: |
NLP7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 959 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEP]
|
| Biological Process: |
stomatal movement
[IMP]
response to nitrate [IMP] response to water deprivation [IMP] nitrate assimilation [IMP] |
| Molecular Function: |
transcription factor activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCEPDDNSAR NGVTTQPSRS RELLMDVDDL DLDGSWPLDQ IPYLSSSNRM ISPIFVSSSS 60
61 EQPCSPLWAF SDGGGNGFHH ATSGGDDEKI SSVSGVPSFR LAEYPLFLPY SSPSAAENTT 120
121 EKHNSFQFPS PLMSLVPPEN TDNYCVIKER MTQALRYFKE STEQHVLAQV WAPVRKNGRD 180
181 LLTTLGQPFV LNPNGNGLNQ YRMISLTYMF SVDSESDVEL GLPGRVFRQK LPEWTPNVQY 240
241 YSSKEFSRLD HALHYNVRGT LALPVFNPSG QSCIGVVELI MTSEKIHYAP EVDKVCKALE 300
301 AVNLKSSEIL DHQTTQICNE SRQNALAEIL EVLTVVCETH NLPLAQTWVP CQHGSVLANG 360
361 GGLKKNCTSF DGSCMGQICM STTDMACYVV DAHVWGFRDA CLEHHLQKGQ GVAGRAFLNG 420
421 GSCFCRDITK FCKTQYPLVH YALMFKLTTC FAISLQSSYT GDDSYILEFF LPSSITDDQE 480
481 QDLLLGSILV TMKEHFQSLR VASGVDFGED DDKLSFEIIQ ALPDKKVHSK IESIRVPFSG 540
541 FKSNATETML IPQPVVQSSD PVNEKINVAT VNGVVKEKKK TEKKRGKTEK TISLDVLQQY 600
601 FTGSLKDAAK SLGVCPTTMK RICRQHGISR WPSRKIKKVN RSITKLKRVI ESVQGTDGGL 660
661 DLTSMAVSSI PWTHGQTSAQ PLNSPNGSKP PELPNTNNSP NHWSSDHSPN EPNGSPELPP 720
721 SNGHKRSRTV DESAGTPTSH GSCDGNQLDE PKVPNQDPLF TVGGSPGLLF PPYSRDHDVS 780
781 AASFAMPNRL LGSIDHFRGM LIEDAGSSKD LRNLCPTAAF DDKFQDTNWM NNDNNSNNNL 840
841 YAPPKEEAIA NVACEPSGSE MRTVTIKASY KDDIIRFRIS SGSGIMELKD EVAKRLKVDA 900
901 GTFDIKYLDD DNEWVLIACD ADLQECLEIP RSSRTKIVRL LVHDVTTNLG SSCESTGEL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
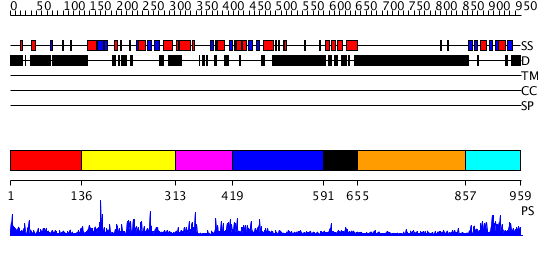
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [136..312] | 2.082967 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [313..418] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [419..590] | 4.141994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [591..654] | 28.69897 | No description for PF02042.7 was found. No confident structure predictions are available. | |
| 6 | View Details | [655..856] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [857..959] | 20.0 | Solution Structure of the PB1 domain of PKCiota |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)