













| General Information: |
|
| Name(s) found: |
gi|9758827
[NCBI NR]
gi|15238474 [NCBI NR] gi|124300972 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 268 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYATSSILSP TPQSFFLSHH LPPISFLYRI NFLGFPVTSC CYGGDIGLAS LYKRRSSIQR 60
61 RRNRIFVTRA RSSPYEILGV SPSATPQDIK RAYRKLALKY HPDVNKEANA QEKFLKIKHA 120
121 YTTLINSDSR RKYGSDSRAT GSSTGQTSRK GNSQVEEDFY GLGDFFKDLQ EEYKNWEASA 180
181 SSQGKPKSLW EELSEIGEEF VEFLEKELNI SDEDNEGSSK NGERFDFEEG STEKSSGKNN 240
241 SSTKNSIEDN IDEIEATLAQ LKKDLGLQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
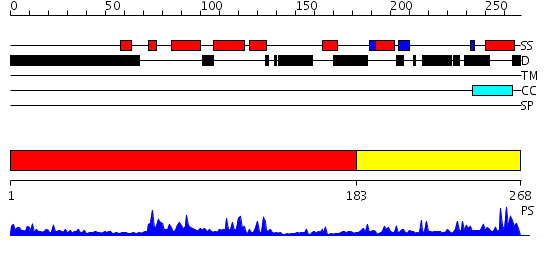
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..182] | 24.0 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [183..268] | 15.0 | The crystal structure of Hsp40 Ydj1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.36 |
Source: Reynolds et al. (2008)