













| General Information: |
|
| Name(s) found: |
SCAR3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1020 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SCAR complex
[TAS]
|
| Biological Process: |
positive regulation of actin nucleation
[IMP]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPRNVYGMNQ SEVYRNVDRE DPKAILNGVA VTGLVGVLRQ LGDLAEFAAE IFHGIQEEVM 60
61 ATASRSNQLK IRLQHIEATV PPLEKAMLAQ TTHIHFAYTG GLEWHPRIPI TQNHLIYDDL 120
121 PHIIMDPYEE CRGPPRLHLL DKFDINGPGS CLKRYSDPTY FRRASSNLSQ GNKKFQKDKK 180
181 HCKMKKKKTS SRSRDMSRLA SLANQNARKT FASFSFSGQT SSTKTTSTSD MEKRYDFQDH 240
241 HSRSFESRSG SGYNECLSTA TSSLKTGERP KGVFVSSSLT PGSCTIASVL SECETEDAHD 300
301 NFQFSPSQGQ AARGSSCVSW DEKAEIVESL GLQTDEASEM VEANSVVDTL DEKPSYGEGI 360
361 GGVDFHSKDN ENDKSESGLR KRAGIDEVRE IKNGREIVGE PRDSEQETES EGECFVDALN 420
421 TIESESENNQ GLQTSQVSSS CGVADERLEK SVCEQETEQN SYSVEDSCRS MDGLMANSFK 480
481 NEENASSENV SVEMHQQNLQ AGSDINRLQK NDLCANKDMR NDSGGKDTIT FTFVPGLENS 540
541 LVDSSNPLIH HGLQENQETE AESSGDLEAF KIWTNGGLLG LKPSKPPVLA MPSSLSPDCK 600
601 TEERTVGFAE AEKDKADDLV ENASHRHVLN NSSLATPGTQ NPGSSNGIVM GIVDQRESHE 660
661 TSSGVFGLSH KFLTSGFRRK DSFAHDRKTV PATIPENDEV TTERRRFCDQ DINEKTFMDP 720
721 FRDEAPIDWI TSSPPLQHMK ISLNPADTLQ ASRLKLKFSD GDNTYNTFSS FQLLPETGTS 780
781 LPDSYSDDDT FCRSSPYMSD TDYLSDNHSL SNSEPWEESS DSHGRKEQEL YDSFHESRHV 840
841 DNNAEASPLG IKSESSCVAV NLSYLQNPAE PLPPPFPPMQ WMVSKTPSEK MEDKTQSLQL 900
901 QEALRFAFEK HISLPTAKNE LPSMVTSAPK PEIKAHLKNN VREEKQSANA KETETGDFLQ 960
961 QIRTQQFNLR PVVMTTTSSA TATTDPIINT KISAILEKAN SIRQAVASKD GDESDTWSDT1020
1021 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
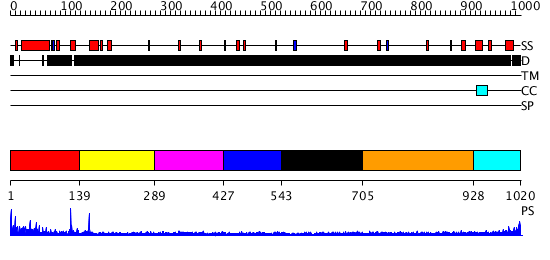
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | 1.499975 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [139..288] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [289..426] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [427..542] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [543..704] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [705..927] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [928..1020] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)