













| General Information: |
|
| Name(s) found: |
gi|4056454
[NCBI NR]
gi|25511660 [NCBI NR] gi|15220851 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 895 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
plant-type cell wall organization
[IEA]
|
| Molecular Function: |
structural constituent of cell wall
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVAASYEPYT YSSPPPPLYD SPTPKVDYKS PPPPYVYSSP PPPLSYSPSP KVDYKSPPPP 60
61 YVYSSPPPPY YSPSPKVEYK SPPPPYVYSS PPPPYYSPSP KVDYKSPPPP YVYSSPPPPY 120
121 YSPSPKPTYK SPPPPYVYNS PPPPYYSPSP KVEYKSPPPP YVYSSPPPPY YSPSPKVDYK 180
181 SPPPPYVYNS PPPPYYSPSP KPTYKSPPPP YIYSSPPPPY YSPSPKPVYK SPPPPYVYSS 240
241 PPPPYYSPSP KPAYKSPPPP YVYSSPPPPY YSPSPKPIYK SPPPPYVYNS PPPPYYSPSP 300
301 KPAYKSPPPP YVYSFPPPPY YSPSPKPVYK SPPPPYVYNS PPPPYYSPSP KPAYKSPPPP 360
361 YVYSSPPPPY YSPSPKPTYK SPPPPYVYSS PPPPYYSPSP KPVYKSPPPP YIYNSPPPPY 420
421 YSPSPKPSYK SPPPPYVYSS PPPPYYSPSP KLTYKSSPPP YVYSSPPPPY YSPSPKVVYK 480
481 SPPPPYVYSS PPPPYYSPSP KPSYKSPPPP YVYNSPPPPY YSPSPKVIYK SPPHPHVCVC 540
541 PPPPPCYSHS PKIEYKSPPT PYVYHSPPPP PYYSPSPKPA YKSSPPPYVY SSPPPPYYSP 600
601 APKPVYKSPP PPYVYNSPPP PYYSPSPKPT YKSPPPPYVY SSPPPPYYSP TPKPTYKSPP 660
661 PPYVYSSPPP PYYSPSPKPT YKSPPPPYVY SSPPPPYYSP APKPTYKSPP PPYVYSSPPP 720
721 PYYSPSPKPT YKSPPPPYVY SSPPPPPYYS PSPKVEYKSP PPPYVYSSPP PPYYSPSPKV 780
781 EYKSPPPPYV YSSPPPPPYY SPSPKVEYKS PPPPYVYSSP PPPTYYSPSP KVEYKSPPPP 840
841 YVYNSPPPPA YYSPSPKIEY KSPPPPYVYS SPPPPSYSPS PKAEYKSPPP PSLYY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
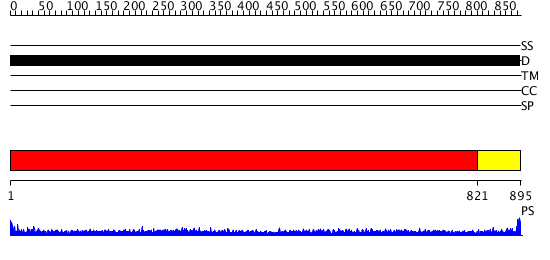
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..820] | 9.522879 | No description for 1y0fA was found. | |
| 2 | View Details | [821..895] | 8.60206 | No description for PF04554.5 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)