













| General Information: |
|
| Name(s) found: |
PUM3_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 964 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
RNA binding
[IEA][ISS]
binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMIPELGRRP MHRGNEDSSF GDDYEKEIGV LLGEQQRRQV EADELERELN LYRSGSAPPT 60
61 VDGSLSAAGG LFSGGGGASF LEFGGVNKGN GFGGDDEEFR KDPAYLSYYY ANMKLNPRLP 120
121 PPLMSREDLR VAQRLKGSNN VLGGVGDRRK VNDSRSLFSM PPGFEAGKPG ASASEWDANG 180
181 LIGLGLGGKQ KSFADIFQAD MGHPVVQQPS RPASRNTFDE NVDSNNNLSP SASQGIGAPS 240
241 PYCYAAVLGS SLSRNGTPDP QGIARVPSPC LTPIGSGRVS SNDKRNTSNQ SPFNGVTSGL 300
301 NESSDLVNAL SGLNLSGTGG LDERGQAEQD VEKVRNYMFG LQDGHNEVNP HGFPNRSDQA 360
361 RGTASCRNSQ MRGSQGSAYN SGSGVANPYQ HHDSPNYYAL NPAVASMMAN QLGTNNYSPM 420
421 YENASATLGY SAMDSRLHGG SFVSSGQNLS ESRNIGRVGN RMMEGGTGHP SHLADPMYHQ 480
481 YARFSENADS FDLLNDPSMD RNYGNSYMNM LEIQRAYLGS QKSQYGLPYK SGSPNSHSYY 540
541 GSPTFGSNMS YPGSPLAHHG MPNSLMSPYS PMRRGEVNMR YPAATRNYTG GVMGSWHMDA 600
601 SLDEGFGSSM LEEFKSNKTR GFELSEIAGH VVEFSSDQYG SRFIQQKLET ATTDEKNMVY 660
661 EEIMPKALAL MTDVFGNYVI QKFFEHGLPP QRRELGEKLI DNVLPLSLQM YGCRVIQKAI 720
721 EVVDLDQKIQ MVKELDGHVM RCVRDQNGNH VVQKCIECVP EENIEFIIST FFGHVVTLST 780
781 HPYGCRVIQR VLEHCHNPDT QSKVMEEILS TVSMLTQDQY GNYVVQHVLE HGKPDERTVI 840
841 IKELAGKIVQ MSQQKFASNV VEKCLTFGGP EERELLVNEM LGTTDENEPL QAMMKDQFAN 900
901 YVVQKVLETC DDQQRELILT RIKVHLNALK KYTYGKHIVA RVEKLVAAGE RRMALQSLPQ 960
961 PLVA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
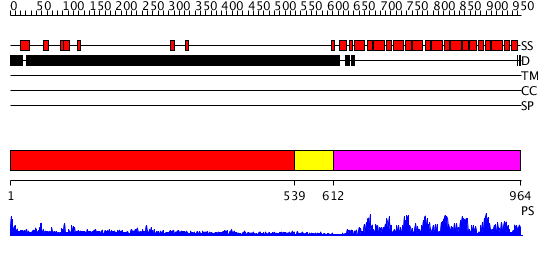
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..538] | 13.0 | No description for 1y0fA was found. | |
| 2 | View Details | [539..611] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [612..964] | 145.0 | Pumilio 1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)