













| General Information: |
|
| Name(s) found: |
MTA70_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 685 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
nuclear speck [IDA] |
| Biological Process: |
embryonic development ending in seed dormancy
[NAS]
mRNA methylation [IMP] |
| Molecular Function: |
methyltransferase activity
[IEA]
S-adenosylmethionine-dependent methyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METESDDATI TVVKDMRVRL ENRIRTQHDA HLDLLSSLQS IVPDIVPSLD LSLKLISSFT 60
61 NRPFVATPPL PEPKVEKKHH PIVKLGTQLQ QLHGHDSKSM LVDSNQRDAE ADGSSGSPMA 120
121 LVRAMVAECL LQRVPFSPTD SSTVLRKLEN DQNARPAEKA ALRDLGGECG PILAVETALK 180
181 SMAEENGSVE LEEFEVSGKP RIMVLAIDRT RLLKELPESF QGNNESNRVV ETPNSIENAT 240
241 VSGGGFGVSG SGNFPRPEMW GGDPNMGFRP MMNAPRGMQM MGMHHPMGIM GRPPPFPLPL 300
301 PLPVPSNQKL RSEEEDLKDV EALLSKKSFK EKQQSRTGEE LLDLIHRPTA KEAATAAKFK 360
361 SKGGSQVKYY CRYLTKEDCR LQSGSHIACN KRHFRRLIAS HTDVSLGDCS FLDTCRHMKT 420
421 CKYVHYELDM ADAMMAGPDK ALKPLRADYC SEAELGEAQW INCDIRSFRM DILGTFGVVM 480
481 ADPPWDIHME LPYGTMADDE MRTLNVPSLQ TDGLIFLWVT GRAMELGREC LELWGYKRVE 540
541 EIIWVKTNQL QRIIRTGRTG HWLNHSKEHC LVGIKGNPEV NRNIDTDVIV AEVRETSRKP 600
601 DEMYAMLERI MPRARKLELF ARMHNAHAGW LSLGNQLNGV RLINEGLRAR FKASYPEIDV 660
661 QPPSPPRASA METDNEPMAI DSITA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
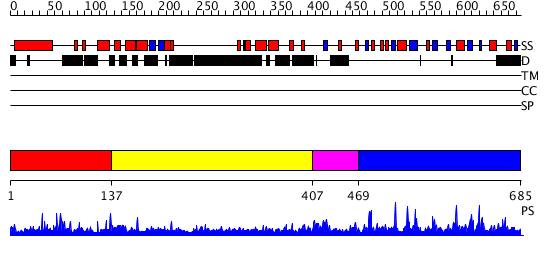
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [137..406] | 1.051996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [407..468] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [469..685] | 92.769551 | No description for PF05063.5 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)