













| General Information: |
|
| Name(s) found: |
HDG7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 682 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA][ISS]
regulation of transcription [IEA] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] sequence-specific DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNGDLEVDMS RGDFNPSFFL GKLKDDEFES RSLSDDSFDA MSGDEDKQEQ RPKKKKRKTK 60
61 YHRHTSYQIQ ELESFFKECP HPNEKQRLEL GKKLTLESKQ IKFWFQNRRT QMKTQLERHE 120
121 NVILKQENEK LRLENSFLKE SMRGSLCIDC GGAVIPGEVS FEQHQLRIEN AKLKEELDRI 180
181 CALANRFIGG SISLEQPSNG GIGSQHLPIG HCVSGGTSLM FMDLAMEAMD ELLKLAELET 240
241 SLWSSKSEKG SMNHFPGSRE TGLVLINSLA LVETLMDTNK WAEMFECIVA VASTLEVISN 300
301 GSDGSRNGSI LLMQAEFQVM SPLVPIKQKK FLRYCKQHGD GLWAVVDVSY DINRGNENLK 360
361 SYGGSKMFPS GCIIQDIGNG CSKVTWIEHS EYEESHTHSL YQPLLSSSVG LGATKWLATL 420
421 QRQCESFTML LSSEDHTGLS HAGTKSILKL AQRMKLNFYS GITASCIHKW EKLLAENVGQ 480
481 DTRILTRKSL EPSGIVLSAA TSLWLPVTQQ RLFEFLCDGK CRNQWDILSN GASMENTLLV 540
541 PKGQQEGSCV SLLRAAGNDQ NESSMLILQE TWNDVSGALV VYAPVDIPSM NTVMSGGDSA 600
601 YVALLPSGFS ILPDGSSSSS DQFDTDGGLV NQESKGCLLT VGFQILVNSL PTAKLNVESV 660
661 ETVNNLIACT IHKIRAALRI PA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
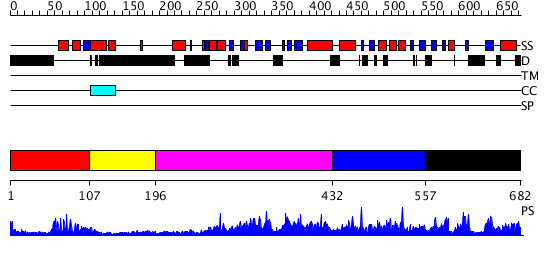
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 21.39794 | Paired protein | |
| 2 | View Details | [107..195] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [196..431] | 3.48 | No description for 2r55A was found. | |
| 4 | View Details | [432..556] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [557..682] | 2.059972 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)