













| General Information: |
|
| Name(s) found: |
PUM1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 968 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
RNA binding
[IEA][ISS]
binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIPELGRRPM HRGNEDSSFG DDYEKEIGVL LGEQQRRQVE ADELERELNL FRSGSAPPTV 60
61 DGSVSAAGGL FSGGGGAPFL EFGGVNKGNG FGGDDEEFRK DPAYLSYYYA NMKLNPRLPP 120
121 PLMSREDLRV AQRLKGSNNV LGGVGDRRKV NDNRSLFSMP PGFEGEKTGA SASEWDANGL 180
181 IGLPGLGLGG KQKSFADIFQ ADMGHGHPVA QQPSRPASRN TFDENVDSNN NLSPSASQGI 240
241 GAPSPYSYAA VLGSSLSRNG TPDPQAIARV PSPCLTPIGS GRMSSNDKRN TSNQSPFNGV 300
301 TSGLNESSDL VNALSGLNLS CSVGLDDRSQ AEQDVEKVRN YMFGLQGGHN EVNQHEFPNK 360
361 SDQAHKATGS LRNSQLRGPH GSAYNGGVGL ANPYQQLDSP NYCLNNYALN PAVASMMANQ 420
421 LGNNNFAPMY DNVSALGFSG MDSRHHGRGF VSSGQNLSES RNLGRFSNRM MGGGAGLQSH 480
481 MVDPMYNQYA DSLDLLNDPS MDRNFMGGSS YMDMLELQRA YLGAQKSQYG VPYKSGSPNS 540
541 HSYYGSPTFG SNMSYPGSPL AHHGMPNSLM SPYSPMRRDE VNMRFPSATR NYSGGLMGSW 600
601 HMDASFDEGF GSSMLEEFKS NKTRGFELSE IAGHVVEFSS DQYGSRFIQQ KLETATTDEK 660
661 NMVYEEIMPQ ALVLMTDVFG NYVIQKFFEH GLPPQRRELA EKLFDHVLPL SLQMYGCRVI 720
721 QKAIEVVDLD QKIKMVKELD GHVMRCVRDQ NGNHVVQKCI ECVPEENIEF IISTFFGHVV 780
781 TLSTHPYGCR VIQRVLEHCH DPDTQSKVME EILSTVSMLA QDQYGNYVVQ HVLEHGKPDE 840
841 RTVIIKELAG KIVQMSQQKF ASNVVEKCLT FGGPEERELL VNEMLGTTDE NEPLQAMMKD 900
901 QFANYVVQKV LETCDDQQRE LILTRIKVHL TALKKYTYGK HVVARIEKLV AAGERRMALQ 960
961 SLTQPQMA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
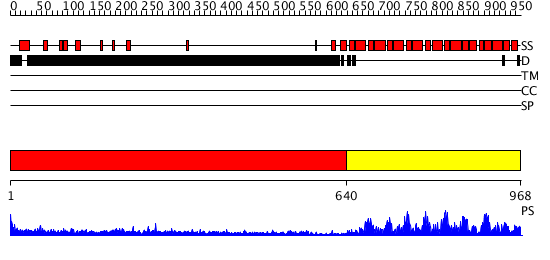
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..639] | 23.09691 | No description for 1y0fB was found. | |
| 2 | View Details | [640..968] | 147.0 | Pumilio 1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)