













| General Information: |
|
| Name(s) found: |
gi|7486226
[NCBI NR]
gi|7269894 [NCBI NR] gi|4914419 [NCBI NR] gi|30688502 [NCBI NR] gi|20465437 [NCBI NR] gi|18377650 [NCBI NR] |
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 828 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
regulation of Rab GTPase activity
[IEA]
|
| Molecular Function: |
Rab GTPase activator activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVPSEIESLG DESDRRFANL RGLRWRVNLG VLPFQSSSID DLRKATAESR RRYAALRRRL 60
61 LIDPHLSKDV RNSPDLSIDN PLSQNPDSTW GRFFRNAELE KTLDQDLSRL YPEHWSYFQA 120
121 PGCQGMLRRI LLLWCLKHPE YGYRQGMHEL LAPLLYVLHV DVDRLSEVRK SYEDHFIDRF 180
181 DGLSFEERDI TYNFEFKKFL EDFTDDEIGG IQGNSKKIKS LDELDPEIQS IVRLSDAYGA 240
241 EGELGIVLSE KFMEHDAYCM FDALMNGVHG CVAMAGFFAY SPASGSHTGL PPVLEASTAF 300
301 YHLLSFVDSS LHSHLVELGV EPQYFGLRWL RVLFGREFLL QDLLIVWDEI FSADNTTRTD 360
361 ADNNTNQSYK IFDSPRGALI SGMAVSMILC LRSSLLATEN AASCLQRLLN FPEKIDVRKI 420
421 IEKAKSLQTL ALDDDVRSSA LSINDGFDQS ISPAVPARTN SFPSGSTSPK SPLIITPQSY 480
481 WEDQWRVLHK AAEEEKKSPS PIQKKKPWFR VKRLFRAESE PTHSAKSPNG KSEVKISSVA 540
541 RNLLEDFNRQ LVSEPVEANP IDVVNNEDSS IRETEDINTD FETAAEESIV MEENSSDLFS 600
601 DPNSPLRDSN YIENDSDSSN ESNLFPDETV KDRETSVVDS PLSISSQPSM EFIVSLSKDQ 660
661 ETSVVDSPLP VSSQPSIEFP VTQSNDEDNA ADKSVANIKE RSKVLPGKFQ WFWKFGRNVT 720
721 AEETRCNGVE SSKSDLVCSS ESHSLPQASS SGSKGDTDQN VMNTLKNLGN SMLEHIQVIE 780
781 SVFQQERGQV QAGLIENLSK NNLVEKGQVT AMTALKELRK ISNLLLEM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
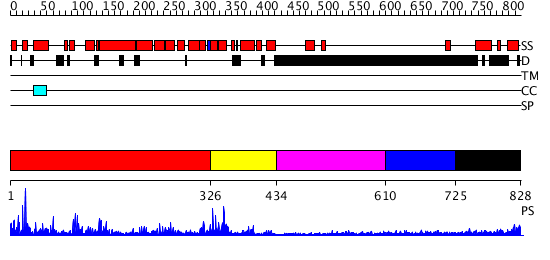
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..325] | 41.69897 | No description for 3dzxA was found. | |
| 2 | View Details | [326..433] | 36.09691 | No description for 2qq8A was found. | |
| 3 | View Details | [434..609] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [610..724] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [725..828] | 1.132988 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)