













| General Information: |
|
| Name(s) found: |
DCE5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 494 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
glutamate metabolic process
[IEA]
carboxylic acid metabolic process [IEA] |
| Molecular Function: |
calmodulin binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLATNSDSD EHLHSTFASR YVRAVVPRFK MPDHCMPKDA AYQVINDELM LDGNPRLNLA 60
61 SFVTTWMEPE CDKLIMDSVN KNYVDMDEYP VTTELQNRCV NMIANLFHAP VGEDEAAIGC 120
121 GTVGSSEAIM LAGLAFKRKW QHRRKAQGLP IDKPNIVTGA NVQVCWEKFA RYFEVELKEV 180
181 KLSEDYYVMD PAKAVEMVDE NTICVAAILG STLTGEFEDV KQLNDLLAEK NAETGWETPI 240
241 HVDAASGGFI APFLYPDLEW DFRLPWVKSI NVSGHKYGLV YAGVGWVVWR TKDDLPEELV 300
301 FHINYLGADQ PTFTLNFSKG SSQIIAQYYQ FIRLGFEGYK NIMENCMDNA RRLREGIEMT 360
361 GKFNIVSKDI GVPLVAFSLK DSSKHTVFEI AESLRKFGWI IPAYTMPADA QHIAVLRVVI 420
421 REDFSRGLAD RLITHIIQVL KEIEGLPSRI AHLAAAAAVS GDDEEVKVKT AKMSLEDITK 480
481 YWKRLVEHKR NIVC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
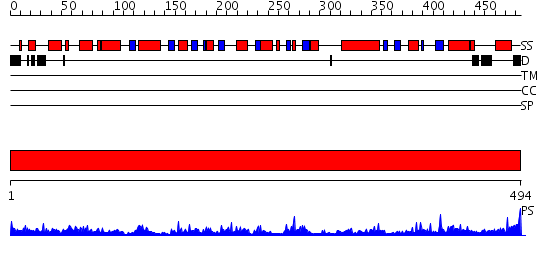
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..494] | 74.09691 | Crystal structure of Escherichia coli GadB (low pH) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)