













| General Information: |
|
| Name(s) found: |
IDE1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 970 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
proteolysis
[IEA][ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
catalytic activity [IEA] metal ion binding [IEA] metalloendopeptidase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVEKSNTTV GGVEILKPRT DNREYRMIVL KNLLQVLLIS DPDTDKCAAS MSVSVGSFSD 60
61 PQGLEGLAHF LEHMLFYASE KYPEEDSYSK YITEHGGSTN AYTASEETNY HFDVNADCFD 120
121 EALDRFAQFF IKPLMSADAT MREIKAVDSE NQKNLLSDGW RIRQLQKHLS KEDHPYHKFS 180
181 TGNMDTLHVR PQAKGVDTRS ELIKFYEEHY SANIMHLVVY GKESLDKIQD LVERMFQEIQ 240
241 NTNKVVPRFP GQPCTADHLQ ILVKAIPIKQ GHKLGVSWPV TPSIHHYDEA PSQYLGHLIG 300
301 HEGEGSLFHA LKTLGWATGL SAGEGEWTLD YSFFKVSIDL TDAGHEHMQE ILGLLFNYIQ 360
361 LLQQTGVCQW IFDELSAICE TKFHYQDKIP PMSYIVDIAS NMQIYPTKDW LVGSSLPTKF 420
421 NPAIVQKVVD ELSPSNFRIF WESQKFEGQT DKAEPWYNTA YSLEKITSST IQEWVQSAPD 480
481 VHLHLPAPNV FIPTDLSLKD ADDKETVPVL LRKTPFSRLW YKPDTMFSKP KAYVKMDFNC 540
541 PLAVSSPDAA VLTDIFTRLL MDYLNEYAYY AQVAGLYYGV SLSDNGFELT LLGYNHKLRI 600
601 LLETVVGKIA NFEVKPDRFA VIKETVTKEY QNYKFRQPYH QAMYYCSLIL QDQTWPWTEE 660
661 LDVLSHLEAE DVAKFVPMLL SRTFIECYIA GNVENNEAES MVKHIEDVLF NDPKPICRPL 720
721 FPSQHLTNRV VKLGEGMKYF YHQDGSNPSD ENSALVHYIQ VHRDDFSMNI KLQLFGLVAK 780
781 QATFHQLRTV EQLGYITALA QRNDSGIYGV QFIIQSSVKG PGHIDSRVES LLKNFESKLY 840
841 EMSNEDFKSN VTALIDMKLE KHKNLKEESR FYWREIQSGT LKFNRKEAEV SALKQLQKQE 900
901 LIDFFDEYIK VGAARKKSLS IRVYGSQHLK EMASDKDEVP SPSVEIEDIV GFRKSQPLHG 960
961 SFRGCGQPKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
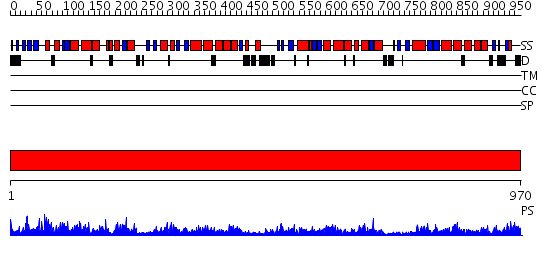
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..970] | 1000.0 | No description for 2g47A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)